













| General Information: |
|
| Name(s) found: |
TEA4_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEIMESHFDP TQQNDSTIIE SRYSPEEYLE QSFEIQRIIS GENSEPQTVA SQEISDSQEE 60
61 DTTLTSSQFE DCGTEYNEVV EDDEFRSEDE DDFMDEEEEY ALYEAELSSS PSIHEEVIDC 120
121 NFVHAIRGFE ATVEGQVDAT KGDMMILLDD SNSYWWLVKM CKNLAIGYLP AEYIETPSER 180
181 LARLNKYKNS ETSNSQQSVT LPPLDIVEKT LEAPSPNFRI KRVTFTCSSN SSDDEMDSEN 240
241 DYEAMVNRTV AENGLEIEFS DSSDSSLSAE YRSESEDHVT DSPAYVDLTE LEGGFNQFNS 300
301 TSFQSTSPLG LEIVETEING SSTTADSKNS HSPYSKFSSA YPDAENSNIS KINISIAGNK 360
361 ELYGNATQSD PSLYSTWIAN KHKTASSATV DSPLRRSLSV DAMQSNASFS SYSSTSNTDK 420
421 SLRPSSYSAV SESSNFTHDV SRDNKEISLN APKSIIVSQS DSFDTSNVTQ DAPNDVEKEP 480
481 ISGQMPNNLS VQSLKQLEVY PIRHSVSIEM PSEKLLSPRL YSSSTPSSPT KGFQKDDEED 540
541 SENRKQADKV ELSPSSLLRQ MSLPVDSSSQ SDAQCTTSSV YITAERKAFS QSSIDLSTLS 600
601 NHHVNNEINR RSFAGGFTSL ADELSEMREL LHESPAPLEC NEEMVIPTPE LDASSAIPSS 660
661 SISHDEDLLP RKNTEESTSS SSFSSLITSP ASLQYDENPF KQSVVAELNN NSSSVPFVDS 720
721 AHASDIHAYD NDHVSTKNKE FNRRLREFIL DPDSLSGLYW SVKSAGVRAS RRVSRNIEGE 780
781 SVSSDLDDIF ANVLKGLSDE MASLLNTNR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
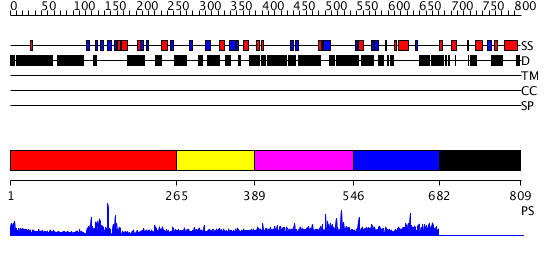
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..264] | 49.154902 | No description for 2h8hA was found. | |
| 2 | View Details | [265..388] | 37.69897 | Hemapoetic cell kinase Hck; Hemopoetic cell kinase Hck; Haemopoetic cell kinase Hck | |
| 3 | View Details | [389..545] | 2.121993 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [546..681] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [682..809] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)