













| General Information: |
|
| Name(s) found: |
YDK2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 757 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MILYIVLPFY VRTKPYSILP SYSSLQWITN AWCSLPSIYI LCHCSKHQEE ITILALKPLI 60
61 AMNNSEEWPL KHVQPPKLSF RHLLSRSVTD APSLRVRWFY AVDRPLRKSR TGPTEIKKAK 120
121 NFLPFSAEDS EHIEKSYLKA VENDGQSEPV NVNEDYLYSV NVVSRELSPI YWDGPVYRIL 180
181 RGTWFFSRGD KLYPCEENLA TQVEEGYLNS CPYREFSNEK DSAAAQSKTW ALLGRYTGGF 240
241 VQYTGSRNAR LVYDDFYRNV SVKIMNRFSP ASFHRSDKLV RGYELDMLES NSKPSTPVPT 300
301 EELTSTTLLN DSSDPSDNFT PSNTESTIDL PSATDASHLM SRPDREVNHL ILCCHGIGQK 360
361 MGERVETVSF VKDISNFRKT LKKTFNSSPD LQAVYPKLKG GGNGVQCLPL LWRQDIRFGM 420
421 ARDLDSSFAD DDDDDDESLN MSRDLALDDL EDDSIPTLDN INIPTVTGLR NIISDVLLDV 480
481 LLYCQPNYRD KILAAVVKRL NRLYNLYKKN VPSFNGHVSL LGHSLGALIL FDIIRYQGNI 540
541 KYSKLQLDFP VANFFALGSP LGLFQMLNGK KIAGPIPKTN LTRSLSYSEQ SFDSGVSILS 600
601 CQNFYNIFHP TDPISYRVEP LVVKQMARLK PQKISHFRPH QDLSSSGVGH KIAGGALNVL 660
661 SGLRSGIANT LILKSLSYAS VFNEATADSH DESQGAENIR DWHIDERMYR LNKTGRIDFM 720
721 LQEGALDTSY SYVSAMNAHS EYWKNVDLAH FILTQLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
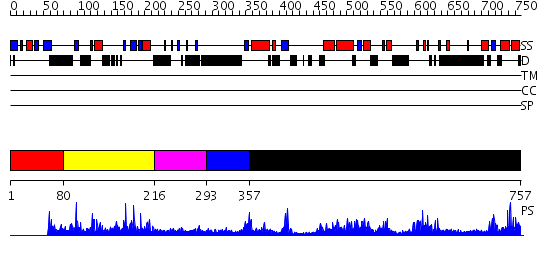
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [80..215] | 2.060988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [216..292] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [293..356] | 1.22 | No description for 2hihA was found. | |
| 5 | View Details | [357..757] | 1.14 | The crystal structure of palmitoyl protein thioesterase-2 reveals the basis for divergent substrate specificities of the two lysosomal thioesterases (PPT1 and PPT2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)