













| General Information: |
|
| Name(s) found: |
CYK3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 886 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIPKQLPCM VRALYAWPGE REGDLKFTEG DLIECLSIGD GKWWIGRHIN TNTQGIFPSN 60
61 FVHCLDIPTV RPGSSMSRTS ASSFRYSSPQ KSSIDTPITS SDQGLTPDLV GSSNALNKPT 120
121 RESDSLKHQK SHPMLNSLGS SLSLKKSVSR PPSSMSRTNL DVSSRWDNTA DNDSQIDAQD 180
181 LSRSTPSPLR SAMENVLQSL NAMGKKSFSR ANSPLLRLTK STTTSKETDF IPVPPAHGST 240
241 SMTSRSKLDD STDNSSKPRT SLQPGESPMK SSRDISRKPS MASSVLSPSD YFPQHRRFQS 300
301 APAPIPRPVS TLIPLTQVRT TASNVIKPRP QTTERPSTAQ SFRKGGGFLK KTFKKLLRRG 360
361 SSKRKPSLQP TPPVSYPAHN VPQKGVRPAS PHTLKSVKDD LKRTKTYTKA EFAAKREEIF 420
421 NKLSMPVYEP LKDLSECIGN VLADGEPVQF SVGTNIHNMN FSAIDKQIRS IIPQRVQVSP 480
481 AVLAKNYLAP GQTTALAQMR AVFIYISERI TFTNQTLDND ELRTSTQVIS EGQGTPFEVA 540
541 LLVKEMLQAL DLWCEVIEGY LKSPDDIYYT RDININHAWN VVTFDNEVRL IDASFASPTH 600
601 PQQALKSSSS NDFYFLMKPN ECIFTHVPEN PDQQFIMPDL SMPIVMALPW VSSVYFTLGL 660
661 KLRKFNTSIL HLNDLEVLQI EFLAPKDIEC VAEVDALSAL APTADVSQCY KYTLTQAFWE 720
721 TPDIRVMRVK AVMPANNRAA VLRIYAGRLG VSSPVRTAPH PMAMSLPFVH HGKNKALEFV 780
781 TRHPVPHCPS VDLYINSPQC GTLHSGVEYK FNVSAYACQP STSISNTRLA IQTPTGNIVR 840
841 LREERSGNGV IFFSLSLTIN ETGEYRALIL AEKIGRWVVY ATWQAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
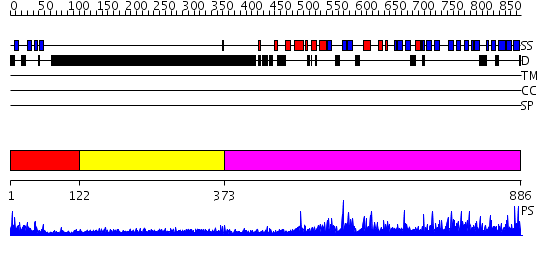
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 7.30103 | No description for 2eyzA was found. | |
| 2 | View Details | [122..372] | 5.314997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [373..886] | 1.42 | No description for 2qshA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)