













| General Information: |
|
| Name(s) found: |
YDY2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1237 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNCRRKPLPL HSNEVQAAES ISKDELYSWT LKNVLILFLV QYRASTPSAL QTSSFSTPVE 60
61 KKTNGAFSGS LFSAKEKKDN PKYPKDFIKY LSLALESISM GKEPEYQDQL IRGCFAAFYN 120
121 NINKSHISKQ LKDSRKIEDM FLIFASSVSS ELTKRLGTAY DSNLADRITA SFIELCKRVT 180
181 LKGQLASPSS EFVSHLDMYH SKLLNKSEIL STKRVNRGQD QFDLSSFPLL KVFSSVFKVA 240
241 YSKALLDAEK LFPSIDEAAI AEDIGILIED LESDACKFSH SVVDFSNYKS YSTWKARELQ 300
301 TLEQHRKIML SLIPLSSSPN HPIQPIGFPA SRKTPTYVPP IARPYFRQLL SLCLEYRLQV 360
361 TDPVEVKQIN TFIRECSIYW RILPSTRCAL SMDFAREHLQ KGLITSADVL THFGELHKSL 420
421 KEWEITYWPK IDVSIFSSAL FGIRNALAQL LKKSLLDLLH EKKADFHRWL NIMSICVEDG 480
481 KVINQDLSPK SEISSVALAL QEHCKVIFDE EFDLSKLPSS NFLDEIILAS NRISVRIKFL 540
541 ERNCSECIYG VLSLPQIFIN IVLPNYIRAA LVVAKEYLRE KANADINDLT KDMLEIYSDL 600
601 KQMIHVLQEY HNEEYRIGSF EGFFQPFIDS WLDNVEASAE QWFLRSLEKD NFDENSSEGQ 660
661 YSSSIVDLFH AFHQAFRTLE GFEWEDDLAN ARFFTRFFRV IYIVLSKYTQ WTTQKFFEEA 720
721 NKQDDTSEVQ NDNSSSWFSK ARNILSGSNE VLPFRFSPTM CILLNNIHFA MHAYEELEQK 780
781 IDLQRLIEAL DVAESTKRHK ISATNYLYTI KVVRGEGLHP DGAGKIRTSY IVLTDNKGRR 840
841 IGKTRPIHSM NPRWDDTFEV KTKDALMITA NLWSKGKFND HEIFGRSSFT LSPKIYGDYL 900
901 PREEWFDLNP HGELLLRIEM EGEREHIGFY VGRTYHDLER AQREMIKFIV NKMEPVINQN 960
961 LSTATLKKLL SANTWMDLDK TMTSVTSLLN RTGFSSKSSE NVKKEGELTD VQIEAAIYEL1020
1021 LDYFDLNFSI IAKHLTKDVF VTVMSYVWDE VVCTTEELLL PPVSVKPINR KPLSPAQMAI1080
1081 VYRWLQFLKD YFYANGEGVK LPVLETDHYK ELLRVQEYYD KPTDFLLQEC DKIASQLYLS1140
1141 SRMVNNEPVE SLPRHSRIKF ANKEIYRSGT IRKVHIQTNE SELERNERII LRILRMRADS1200
1201 KRFLSKHFQR KGRLLMADAM RNGYVMPLGH LKKLDSR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
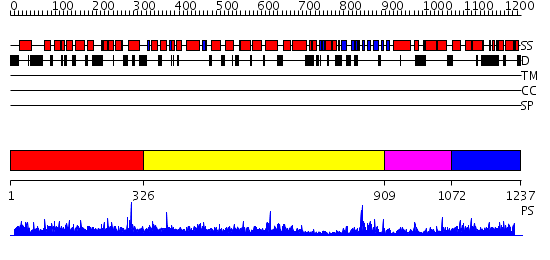
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..325] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [326..908] | 26.0 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) | |
| 3 | View Details | [909..1071] | 46.30103 | Synaptogamin I | |
| 4 | View Details | [1072..1237] | 1.081985 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)