













| General Information: |
|
| Name(s) found: |
YD83_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1004 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFFKKKLSK GKEILSKSNL KTHTSSNASL SIDDLNRFGF SLNPVLWCLD HQQGLLAIVS 60
61 STNRIYIYGK QHVQSVIVPD CSTIVHIALC AAYLIVIDSR NTVLSYPLMK HRDLSKPAAT 120
121 YFLKQKVTCT VTDPTIDWVF FGMSDGSVVP WDVTRHCLGK FKVPNLYVPR HEEWRMMGYS 180
181 YAPVPGKLSP VVSVQIHPKD LGVILIAYPD GVVLYSIRTD EVIRFYELEY APGSTAAVLS 240
241 PHNYRRPIVK GIEWSPWGDH FVSYYTDSTF AFWDVDQEYP VQVRNFVDSN IHTYTPMQRN 300
301 PPKTELEPIR SMRWCCCEDP TVSFILMLGG LPKEAPVKGI SLFSYRNLPA KKDVETFAEF 360
361 FANPNSQRFF PFIDIPPVRD MLVIPSSSPH YNGSHNPKNL LLLSEDNSLS LLDISTGNIS 420
421 NMSLSIPPSL CFLASDFRVI AFQTVTKKVW NQIEDTISVN SHYSCLFGGS PSPGYLKKLD 480
481 ERNLLITSTG LSLSIWDISQ GFMNPSLCVN LDFSSVMRKH LTPSAFITTA SFSTYNPEFS 540
541 CADSFGRVIV CKRKNHKENL PAQLANGIYR LDDTLVLEGT LHAQYYIDLK RGRVTLNQMS 600
601 NIGFVCIGYQ DGGITIIDMR GPHILCNTSI SELGLERRGK PDPDFLTSAE FVVMNPKGSP 660
661 SSIYVVTGTY RGMTLLFRID PSSSGRFSAY FESSRQLDIK NIYKICSLTQ DGQIATATGS 720
721 SLQSVGYPLP QEVFLVYIGD SGISVFNKIN NQVGNLDWRK PVCCRAALVL STVSKHMGSV 780
781 VCVNSDLSVN WYSLPNLREE RKMQLPLDID KNRLKEGDIL GNGDYIFPTL GAHELAFGCV 840
841 LGSGRTLANL APMMLITHNA SHVPPRPSKS LWNWLLGEQS TSAEELDILL GGENRAESKV 900
901 HTLETPKVIS ARPAESVKQP LTPVPSMTSQ SAQSYIPPRR QQQQKGFFAQ INDHLAQRGN 960
961 MLGGIENTMD DLEEMSAEWA NEIKDSLAGT KKDLILSGLK SYIP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
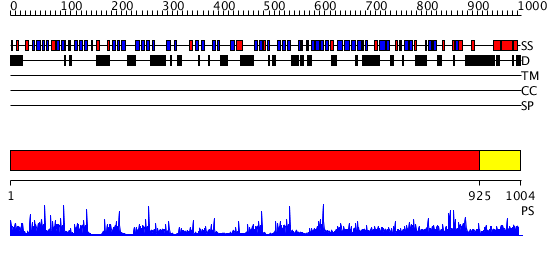
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..924] | 141.0 | No description for 2oajA was found. | |
| 2 | View Details | [925..1004] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)