













| General Information: |
|
| Name(s) found: |
SDA1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 719 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKRRTAAVL PNNLPHLQHL VKKDPKSYRE EFLQQWNHYE TAREIFLVNP SSDISEFCSL 60
61 IDFISQTCNY YHDVTADFPS ELIELLQKNH TIFPFELCEK IVLCLVLLKN KTVISPITLL 120
121 QCFFPLFREN PTRGVRELLY QQVSLTIRNA NKQAKNDKLN KAVQSALFTL VDGSGSGSAT 180
181 HTDYSGQKRE YSASELSGVW AVKMVRDLWK RNVWSGDARA VNIMKSAALS SNPKVMLAGI 240
241 YFFLGADNQE EEEESDDEGP DVSKLLHQAN VNKKTKSREA ALLRARAVVK KKERGKQNVP 300
301 TNVNFPALQL LHDPQGFAEE LFEKHLASSK SRLSMDQKLV VLRLLTRLVG SHKLTVLGLY 360
361 SFLMKYLTPH QRDVTQFLAC LAQASHEFVP PDALEPLVRK IADEFVTSGV ANEVVCAGIN 420
421 AIREVCARAP LAMTPDLLQD LTEYKSSKDK GVMMASRSLI TLYREVAPDM LKRKDRGKLA 480
481 SIEMKDRTPL KYGEELNVTH GIQGLELLAQ YKAEHGEEGE NGDDWDNWEV SEDGQNSDDS 540
541 GGWIDVDSDD NIELSDSDEE EEKATARKES DEKGSSSQKE LVDRMTELAS QSILTPNDLK 600
601 KLEELREQAG VDRLVNGPKR SLKRPDDAVE ADEIEGPRKK AKNDREARIA SVMEGREGRD 660
661 KFSSKKAGFN PTSLSNKRKQ RNKNFMMIKH KLKGKAGRSL VQKQKVLREH VAREKRKIK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
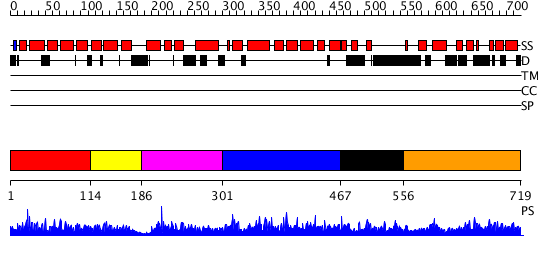
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | 1.021966 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [114..185] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [186..300] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [301..466] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [467..555] | 1.020986 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [556..719] | 1.015981 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)