













| General Information: |
|
| Name(s) found: |
YAS9_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 738 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSSKDSSF QVETPVQNIL ETSTNSELQD QVSSPYEPDY NSPVKQAAAS ISALQTQDDT 60
61 LFNNVDERTL ENKDGNKSDD ANFDQVSGIP SGSLEIPILN SATSNIRLTP SDTYNNIPVS 120
121 DTNNEEISKN IYGAPILEST SSDFQSKDSL STTQPSVSGG NGSTSQSPPS LDVEQNKPFS 180
181 ISNEPVEQET ENSSTKDLQV YDFQTASEHL PEQSLQNTTY YDPSKTYSSV NFEEIEYGKS 240
241 HEKLDLPYRT TDFIPYSKDL STSPEAHRTS IYSYSANLPN YYNEHNELHE HHNPQTPSSP 300
301 ESAYSPENLQ LNHEAQNVEY LGNNAAEKSL QMNLEDEQRF QQFLKDEESI MSNWYPGQFP 360
361 SASRLFLGHL NTKSLSKRNL WKVFKIYGPL AQIVLKANYG FVQFFTNEDC ARALNAEQGN 420
421 FVRGQKLHLE ISKIQKKYQN QIENMKKGSH VTKSNQYSEM IGNLPYPTSS RKRTRSPLMS 480
481 KGKSYDRKGS ISMSKNFSPD CEILVTEDCP KEFVWGVEKV FQERRLNIHT TCLYRDSNLQ 540
541 VIIKSCIINS VKSIILINAG LAHLGKVSVQ VFKDGSSDSE VRCDEYAAVD VMVAASIVHH 600
601 AKTSLMHSAA SSTPSYNGER IVPDVPSPCI STNPNLPALV GSLDSVNLHH LLGFIQNTYS 660
661 TTSYIPTRVS FNPNDTGGSF GTITSQSQFV VNEMPKNYAR DNYEALHSQE SRQRSSVAGN 720
721 KQLQKILEQL AELKQPDF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
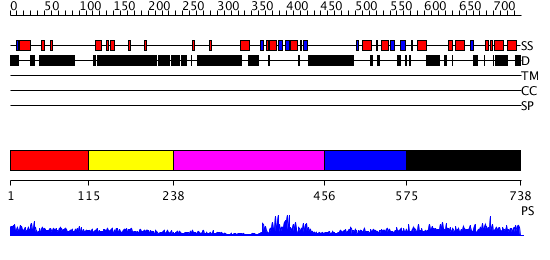
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [115..237] | 1.117996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [238..455] | 22.045757 | Poly(A)-binding protein | |
| 4 | View Details | [456..574] | 25.30103 | Crystal Structure of UP1 Complexed With d(TAGG(6MI)TTAGGG): A Human Telomeric Repeat Containing 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine (6MI) | |
| 5 | View Details | [575..738] | 5.0 | Sec24 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)