













| General Information: |
|
| Name(s) found: |
UBP14_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 775 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSCPHLTETN VVIPDNSQVI YREECVRCFN SQDEEGGIDL CLTCFQSGCG ETGLKHSLVH 60
61 FEQTLHPIVV TIARQPKQKI NDEPPQKITK LEIREDSDED LYDYFYVPKC LVCNIILDIQ 120
121 DPLLSLSLEA MKNATKASNK SQLTAWENEL TTCDHIINLP ENETYVTNLD NATCSKCDLA 180
181 ENLWMCLTCG ALSCGRKQYG GGGGNGHALS HYDDTGHPLA VKLKSISPDG QADIYCYSCD 240
241 EERIDPNIKT HMLNFGIDIA KLNKTEKSLA ELQLEQNLNW DFGASEEDDA SKRLFGPGLT 300
301 GLKNLGNSCY LASTMQSLFS IKEFAIHELN LFNTYNSVCQ TPTTDLQCQL GKLADGLVSG 360
361 KFSKPSKIGL LNNPSSSILP YQDGLRPFMF KDVVGQGHSE FGTSQQQDAY EFLLYLLGKI 420
421 RKSSIAKTDI TKIFDFETEQ KLSCLSCKRV RYSSFSSQGL TLTVPRVKIG EIEGEQIYEE 480
481 VSIDQCLDAT IQPDQMEYTC EACKSKLGAT TTTAMKSFPK VLILQANRFD LQGYQVKKLS 540
541 IPIIVNEDGI YNFDRLMAKD HPNDEDYLPE KTETIEWNQS AIEQLQAMGF PLVRCQRALL 600
601 ATGNSDTETA MNWLFEHMED PEIDKPIEVS ELLPKADSSV SEENVQSLCE FGFTVAQARK 660
661 GLLESNNNIE RAVDWILNHP DESFEEPPLE GSDSSIKNEN MGSWESTNVP VNYNLKAIIS 720
721 HKGSSAHAGH YVAFIRKEID GKQQWVLFND EKVLQVASLE EAKTTGYVYL FERLD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
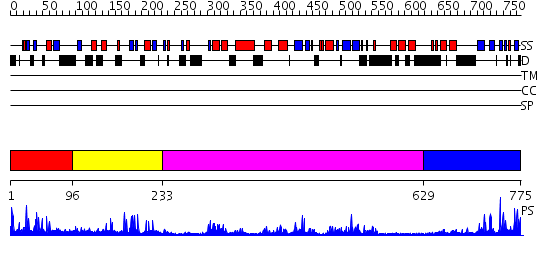
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 14.522879 | No description for 2uzgA was found. | |
| 2 | View Details | [96..232] | 22.69897 | No description for 2g43A was found. | |
| 3 | View Details | [233..628] | 41.0 | Crystal structure of HAUSP | |
| 4 | View Details | [629..775] | 5.16 | Crystal structure of Ubiquitin carboxyl-terminal hydrolase 6 (yfr010w) from Saccharomyces cerevisiae at 1.74 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)