













| General Information: |
|
| Name(s) found: |
YOM2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 749 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTVVGETEL NRLNNGLSSN NGSSADEGLE HWNPLKKIHS ADSSRRRSTS SQLNQPSNYH 60
61 MHSSLHEEVS VGSSIASPSS RIPDKAISQY YETSPPSSTS SLSSNNQLMN SSVILSPGQF 120
121 LPDDANAYGP KASLPPSEMP SHFHPLWSKS ASKDSNAFND PSAIPNPSPL FSSAYATRIN 180
181 GSLRNDKWNR SSFSEALSSS RFSRPQVGQQ QPLSSAFPFQ PVKQPTEQPG SLHPFMQESK 240
241 TSPFATRRPS LNTDHHGRPI LLSPLNYQNS SLNPSTPSPF GGSPVMHPPV SNLSPRTPAV 300
301 PMSSDGHLAP AFDFLNENPI WSKRFSISSI KYSAPSTSNA SNIAPDSAPP ASSQFSVPFN 360
361 AAAENINDTP DSVLANSPTP RHLPYTWSRH STSGPSRSTV LNPSTSRMSN YTGLESHLAQ 420
421 LSFKRRSNSA TLPSLGSIRP FPISEHSPNI NPLDEAAVED EVKEEKSRFH LGHRRSSTAD 480
481 NGTLSSNVPL YPAYNSSPVQ TRTSLFSSRL SKPSNPIVSS VSQANAPKNA LHSMPSPTSL 540
541 ANLPSNLSDT QYKKLYNLYL VEFKAGRADV FYIDDNIKLS LNLNDYVVVD ADRGQDLGRL 600
601 IAQNLSQSEV ASHIEKIPSD RNGQLQNLDG AILEGDTSQS LHPKRILRKA QPHEVDQLIQ 660
661 KTQDEAQALL VCQAKVRQRK LPMEVLDGEY QWDRKKLTFY YHAKQRIDFR ELVRDLFKVY 720
721 KTRIWMCAVS GTNMSSPASI NASTNQFVL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
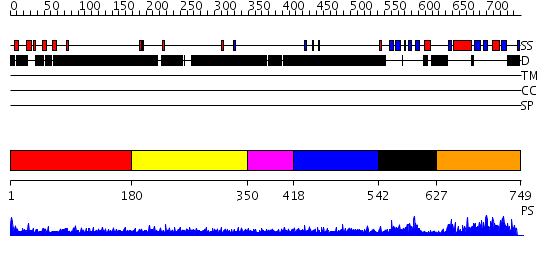
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..179] | 2.209996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [180..349] | 4.224987 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [350..417] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [418..541] | 5.191988 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [542..626] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [627..749] | 50.148742 | No description for PF04468.3 was found. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)