













| General Information: |
|
| Name(s) found: |
T111_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 979 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFDGLIVEN ENTKSGYNDG NDLTDLFKQN GTDMSVINSL LGDTNNPGMN ESPKILDSSF 60
61 ENSNPQDGPN YEDFDFMGSI HKEFGNNINE MDDMEDVSDD NLPEEEQAVN YTGDKDDEDF 120
121 GKLLAKEMGE EAAGQVLSGV GFSIPSGLVP PSEPSKTVSS TTEELQNEAQ IRESIVKTFF 180
181 PTFERGVLLN FSELFKPKPV KLAPPKKKTP KVCVPGRLTL EVDTDYAIIF NSKKSLPLKR 240
241 NVVSPISTHT KKRRRTANTS QRNDGLDLNT VFTTNDWEKN IIYDESDVNK TNQSSFFIDK 300
301 SLVDIDFAFD ENIFDGDTGT SKVVLNLNDP KLLLQPQLPK KEDSQRSFAD THQRNSLAWK 360
361 FNISNDPAYE MLKQNHQSKV RNTLSQLAIE HAAFAEKLTF PYYKTRLSKR AVRSYHRPTM 420
421 SFKPNAAIVF SPLIVRKRSK DKHKSERELI PTTKEITMGD TTHAILVEFS EEHPAVLSNA 480
481 GMASRIVNYY RKKNEQDESR PKLEVGESHV LDVQDRSPFW NFGSVEPGEI TPTLYNKMIR 540
541 APLFKHEVPP TDFILIRNSS SYGSKYYLKN INHMFVSGQT FPVTDVPGPH SRKVTTASKN 600
601 RLKMLVFRLI RRSPNGGLFI RQLSKHFSDQ NEMQIRQRLK EFMEYKKKGD GPGYWKLKSN 660
661 EVVPDEAGTR SMVSPETVCL LESMQVGVRQ LEDAGYGKTM DEINDDEDEE QPAEQLLAPW 720
721 ITTRNFINAT QGKAMLTLFG EGDPTGIGEG YSFIRTSMKG GFKPAGETAD DKPEPQTKNA 780
781 HAYNVAKQQR AYEEEINRIW NAQKRGLSIN NLEELAKKYG INSIHDDYVE SNEETTREET 840
841 PSSDKVLRIV RLYRDKNGNL ERKQETIHDP IVIHAYLKKR REIDEQSTAL DAVVPTGDEA 900
901 IDRRNRRRLE QELAKSQKNW ERRRARHAAK EGINLNGEGR KPTTRKCSNC GQVGHMKTNK 960
961 ICPLFGRPEG GLATMLDKN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
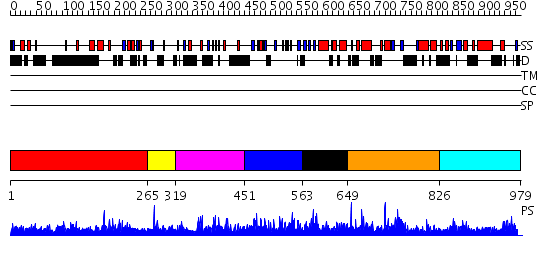
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..264] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [265..318] | 1.026996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [319..450] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [451..562] | 2.03798 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [563..648] | 1.039996 | View MSA. Confident ab initio structure predictions are available. | |
| 6 | View Details | [649..825] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [826..979] | 1.039967 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)