













| General Information: |
|
| Name(s) found: |
NG06_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1315 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDINDPSFSS TENGIYELLS RGYAAIEAAT ERDLKLPELG DIIGQVHAPE YISQVLSGWK 60
61 PFYLRSVVNI PDRIFEQYNR TECFTQMGLF AEIQRAWITV DNRLFLWDYL SGQNFQAYED 120
121 LSHTIVNVKL VRPKANVFVS EIQHLLVIAT SQEMLLLGVT IDEKTGELSF FSTGIQISVQ 180
181 GINVNCIVSS EDGRIFFSGN KDPNLYEFSY QLEEGWFSRR CSKINITGSV FDNFIPSFFS 240
241 FGTHGDGIKQ IAVDDSRSLL YVLRETSSVS CYELTKNGVN RCVFYSFSSM ISQAQMLNAT 300
301 SPLLDPRTTQ IVSIVPIPAY ESQQIYCVAI TSTGCRFYMR GGRGPISHYA PSNSTLSSTP 360
361 PSTLQLTFVR FPPPMQVENY ASSRNYPANP FFLQNQSTSQ QQPERSSAVK TTPMKCSSLS 420
421 NIYTSDLFFA ISSSNTNEGD VVCCTAPEVG RIANAWQSGT QPSLIESSMY VPIKGFVQDI 480
481 KCIQNSRERN ELVSQFNTPP PTFAILTNTG VYVVVHRRPI DVLASAIRMG PSLSSGIDGQ 540
541 VQLFFESVGR AEGCATCLGI VSGCLDQGDF SHAAANFSGS TTKLAQADLL DIVKKYYIEF 600
601 GGKAFIDQSR YNNQYDSSSL EFVRLSGCHD GLASSISRII RNVWKNHVII AKKMQNKRIH 660
661 YAPAFNATEI LKIQSGLLYL STFLENNKSF IEGLNSPNTL IGSSNVADEI AVQAEHRALS 720
721 ALLLVLQQIV EGISFLLFLN DTGVSDFHEI VSSTSIDIQK SCSNMTFGEF FTSKRGREVT 780
781 KELVNSLVNR HLQSGGNIDM VSQLLRKKCG SFCSADDVLI FKAVESLKKA KDTVDIEERQ 840
841 SLIELSYTLF KKAAHVFTPE DLRLAVEEYK SLNAYTTAVN LALHVASARD DRNQALSYLV 900
901 DGMPENDPRR EPFESRTKCY SYIFEILDSL ESQMSNDSSA IKVDVYDTIQ RSKDELFHYC 960
961 FYDWYSFKGL TDRLIEIDSP YIQSYLERNS TKDMKIADLL WQYYAKREQY YQASIVLYDL1020
1021 ATTHLAFSLE QRIEYLTRAK GFGSCHVPNS LRHKMNKVML SVLEQLDVAS IQDDVLIAIR1080
1081 GDMRIPTSKR EELSKQLDGE IIPLSDLFNN YADPLGYGEI CLSIFQCADY RGINEILNCW1140
1141 ESIIKTTHEN AIISPVGSSP VEAVSSTLKN LTLRFSQSEN VFPIEQIIDI TERYAFDQQG1200
1201 EAVATGWVID TFLGAGVSHE LIFIVLNQLY DRREKPWQGK DRLFFLIKEV THLLKLWHEV1260
1261 SVRAGVAQTS KPSFDAPLVL EAIEKYKNAL GAPDTATKSC KENLISLDSE IRQTY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
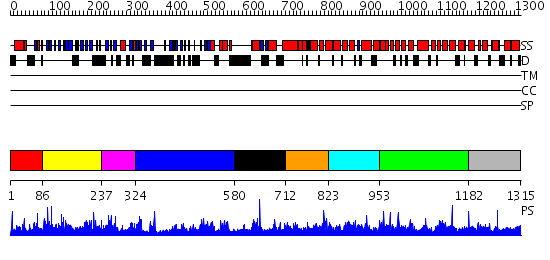
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..236] | 2.041986 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [237..323] | 1.042991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [324..579] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [580..711] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [712..822] | 1.036993 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [823..952] | 1.03897 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [953..1181] | 1.037989 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1182..1315] | 2.038967 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)