













| General Information: |
|
| Name(s) found: |
RHP9_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 778 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEVNDTSLHK GFGLDINSQR VFGAQAAISR NNYSKVNASI NPSPPRSNDN SNKEFSYSKD 60
61 VNNNGAVEEL SLTQLFEVPS QAAFAKQSSQ DISDDELIQH DSRKVISSPY SPKQTHTVLK 120
121 RLYDRQSVIS DHEKLLTPQN VSNSSQILSP FTSLLPSTLS TLKDTPLSVS QNEKNLETVG 180
181 EVLVPETVAQ HRTKFYDYTL DEMENETESG QVETTPTRLA TSLGSPVLYG RVESTPPAFL 240
241 PETSEKQYKR KFSFTEPSSE KVDNTETKFS KKTKNINDEN FPNPFNVISS YETSASPSTV 300
301 IDQSSQVSSI FVNKRLRKSV NNQAISRSDS LSLDTPKIDS LFTRASIKPL KPSQSPNSRR 360
361 SFKNRVLAFF KGYPSFYYPA TLVAPVHSAV TSSIMYKVQF DDATMSTVNS NQIKRFFLKK 420
421 GDVVQSTRLG KIKHTVVKTF RSTNEQLSLI AVDALNNDMV ILAHGEIEVT VPISTIYVAP 480
481 VNIRRFQGRD LSFSTLKDMK FEETSFLPSH DSQRNRSSLK ERDSSFVKKN LDSESNQLIF 540
541 DDCVFAFSGP VHEDAYDRSA LETVVQDHGG LVLDTGLRPL FNDPFKSKQK KLRHLKPQKR 600
601 SKSWNQAFVV SDTFSRKVKY LEALAFNIPC VHPQFIKQCL KMNRVVDFSP YLLASGYSHR 660
661 LDCTLSQRIE PFDTTDSLYD RLLARKGPLF GKKILFIIPE AKSWQKKIEN TEQGQKALAH 720
721 VYHALALGAD VEIRPNVAHL ECDLILTMDG NIVDETNCPV VDPEWIVECL ISQSDIST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
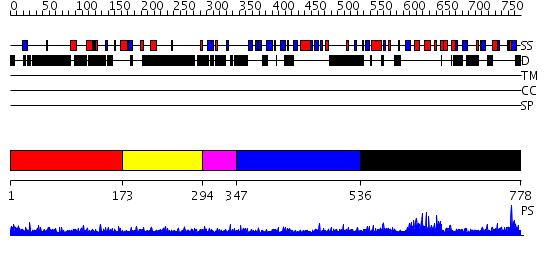
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [173..293] | 1.212992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [294..346] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [347..535] | 5.6 | No description for 2fhdA was found. | |
| 5 | View Details | [536..778] | 69.69897 | 53BP1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)