













| General Information: |
|
| Name(s) found: |
CND2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 742 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRASLGGHA PVSLPSLNDD ALEKKRAKEN SRKQRELRRS SALHSITPRR ESLNNSSPFN 60
61 SSHQVPVLSN FEEWIKLATD NKINSTNTWN FALIDYFHDM SLLRDGEDIN FQKASCTLDG 120
121 CVKIYTSRID SVATETGKLL SGLANDSKVL QQTEEGEDAE NDDEDLQKKK ERKRAQRSVK 180
181 TLVKDFESIR AKKFELECSF DPLFKKMCAD FDEDGAKGLL MNHLCVDQHG RIVFDSSDTV 240
241 IKDLENKDVE AESQEAVVAA PIESHDTEMT NVHDNISRET LNGIYKCYFT DIDQLTICPS 300
301 LQGFEFDSKG NLDVSLLKSL SDEVNMITTT SLVDNTMEKT DADAASLSSD SDGEEGHIVH 360
361 ALEEMAYDEE NPYVDVVPKA MDESENPDFG VDTEVNMADG STMNENYSII STAAANGVYE 420
421 YFDKSMKKNW AGPEHWRIQA LRKNINNAST VFNSSNTAES SDNVSRSLSS TERKKRRELD 480
481 NAIDFLQEVD VEALFTPATS SLKLPKSHWK RHNRCLLPDD YQYDSKRLLQ LFLKPKMSVL 540
541 PNADGEGQLQ LNKALDDEND LDGIQPHGFD SDGSDNVDEG IPPYGFGDSD SPKQTPLLTP 600
601 PSSSGFGDNL LLTARLAKPD MLNYAKRAKK VDVRVLKEKL WKCLDLENTI KENSINSHIE 660
661 GSEMESEETN MPVKSFFSTV NQLEETYEKK ELKDISTSFA FICVLHLANE HNLELTSNED 720
721 FSDVFIRPGP NLTTLEALEN DV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
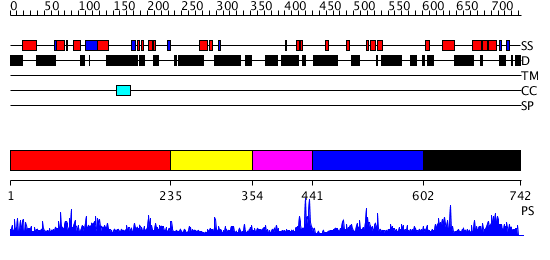
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..234] | 2.021967 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [235..353] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [354..440] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [441..601] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [602..742] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)