













| General Information: |
|
| Name(s) found: |
PLSB_ECOUT
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli UTI89 |
| Length: | 827 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTFCYPCRAF ALLTRGFTSF MSGWPRIYYK LLNLPLSILV KSKSIPADPA PELGLDTSRP 60
61 IMYVLPYNSK ADLLTLRAQC LAHDLPDPLE PLEIDGTLLP RYVFIHGGPR VFTYYTPKEE 120
121 SIKLFHDYLD LHRSNPNLDV QMVPVSVMFG RAPGREKGEV NPPLRMLNGV QKFFAVLWLG 180
181 RDSFVRFSPS VSLRRMADEH GTDKTIAQKL ARVARMHFAR QRLAAVGPRL PARQDLFNKL 240
241 LASRAIAKAV EDEARSKKIS HEKAQQNAIA LMEEIAANFS YEMIRLTDRI LGFTWNRLYQ 300
301 GINVHNAERV RQLAHDGHEL VYVPCHRSHM DYLLLSYVLY HQGLVPPHIA AGINLNFWPA 360
361 GPIFRRLGAF FIRRTFKGNK LYSTVFREYL GELFSRGYSV EYFVEGGRSR TGRLLDPKTG 420
421 TLSMTIQAML RGGTRPITLI PIYIGYEHVM EVGTYAKELR GATKEKESLP QMLRGLSKLR 480
481 NLGQGYVNFG EPMPLMTYLN QHVPDWRESI DPIEAVRPAW LTPTVNNIAA DLMVRINNAG 540
541 AANAMNLCCT ALLASRQRSL TREQLTEQLN CYLDLMRNVP YSTDSTVPSA SASELIDHAL 600
601 QMNKFEVEKD TIGDIIILPR EQAVLMTYYR NNIAHMLVLP SLMAAIVTQH RHISRDVLME 660
661 HVNVLYPMLK AELFLRWDRD ELPDVIDALA NEMQRQGLIT LQDDELHINP AHSRTLQLLA 720
721 AGARETLQRY AITFWLLSAN PSINRGTLEK ESRTVAQRLS VLHGINAPEF FDKAVFSSLV 780
781 LTLRDEGYIS DSGDAEPAET MKVYQLLAEL ITSDVRLTIE SATQGEG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
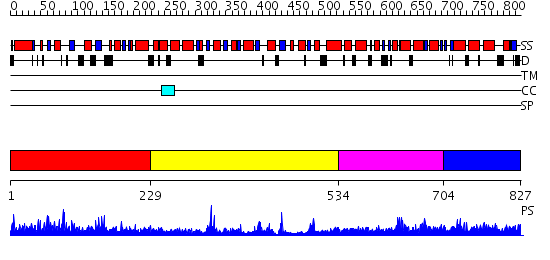
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..228] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [229..533] | 3.522879 | No description for 2q66A was found. | |
| 3 | View Details | [534..703] | 1.20298 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [704..827] | 1.091989 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)