













| General Information: |
|
| Name(s) found: |
gi|168279077
[NCBI NR]
gi|109451850 [NCBI NR] gi|109451272 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 773 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METDESPSPL PCGPAGEAVM ESRARPFQAL PREQSPPPPL QTSSGAEVMD VGSGGDGQSE 60
61 LPAEDPFNFY GASLLSKGSF SKGRLLIDPN CSGHSPRTAR HAPAVRKFSP DLKLLKDVKI 120
121 SVSFTESCRS KDRKVLYTGA ERDVRAECGL LLSPVSGDVH ACPFGGSVGD GVGIGGESAD 180
181 KKDEENELDQ EKRVEYAVLD ELEDFTDNLE LDEEGAGGFT AKAIVQRDRV DEEALNFPYE 240
241 DDFDNDVDAL LEEGLCAPKK RRTEEKYGGD SDHPSDGETS VQPMMTKIKT VLKSRGRPPT 300
301 EPLPDGWIMT FHNSGVPVYL HRESRVVTWS RPYFLGTGSI RKHDPPLSSI PCLHYKKMKD 360
361 NEEREQSSDL TPSGDVSPVK PLSRSAELEF PLDEPDSMGA DPGPPDEKDP LGAEAAPGAL 420
421 GQVKAKVEVC KDESVDLEEF RSYLEKRFDF EQVTVKKFRT WAERRQFNRE MKRKQAESER 480
481 PILPANQKLI TLSVQDAPTK KEFVINPNGK SEVCILHEYM QRVLKVRPVY NFFECENPSE 540
541 PFGASVTIDG VTYGSGTASS KKLAKNKAAR ATLEILIPDF VKQTSEEKPK DSEELEYFNH 600
601 ISIEDSRVYE LTSKAGLLSP YQILHECLKR NHGMGDTSIK FEVVPGKNQK SEYVMACGKH 660
661 TVRGWCKNKR VGKQLASQKI LQLLHPHVKN WGSLLRMYGR ESSKMVKQET SDKSVIELQQ 720
721 YAKKNKPNLH ILSKLQEEMK RLAEEREETR KKPKMSIVAS AQPGGEPLCT VDV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
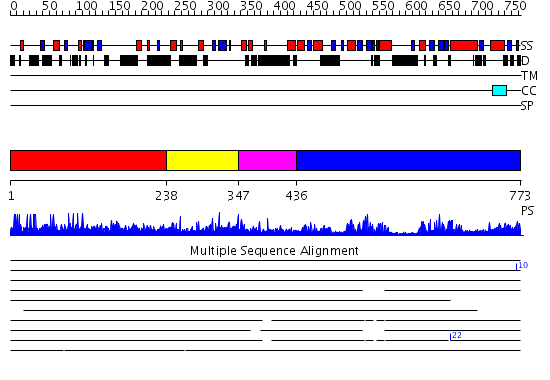
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..237] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [238..346] | 1.035978 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [347..435] | 2.16699 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [436..773] | 50.154902 | No description for 2yt4A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)