













| General Information: |
|
| Name(s) found: |
FBpp0292097
[FlyBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transport vesicle
[ISS]
|
| Biological Process: |
locomotory behavior
[IMP]
glutamine metabolic process [IEA] memory [IMP] |
| Molecular Function: |
carbon-monoxide oxygenase activity
[ISS]
glutaminase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQTTEMEAL KRQSEDGDTK EREENYRRGK EQETTETAAI IEEIIEGISL QNRRGTIVRT 60
61 ISAIISHREQ EQRNAEDVLF DMFASEETGL ISMGKFLAGL KTTGIRRNDP RVRELMDNLK 120
121 KVHKLNNYET GSSAETQHLN RETFKAVVAP NIVLIAKAFR QQFVIPDFTS FVKDIEDIYN 180
181 RCKTNTLGKL ADYIPQLARY NPDSWGLSIC TIDGQRFSIG DVDVPFTLQS CSKPLTYAIA 240
241 LEKLGPKVVH SYVGQEPSGR NFNELVLDQN KKPHNPMINA GAILTCSLMN ALVKPDMTSA 300
301 EIFDYTMSWF KRLSGGEYIG FNNAVFLSER EAADRNYALG FYMRENKCFP KRTNLKEVMD 360
361 FYFQCCSMET NCEAMSVIAA SLANGGICPT TEEKVFRPEV IRDVLSIMHS CGTYDYSGQF 420
421 AFKVGLPAKS GVSGGMMLVI PNVMGIFAWS PPLDHLGNTV RGLQFCEELV SMFNFHRYDN 480
481 LKHLSNKKDP RKHRYETKGL SIVNLLFSAA SGDVTALRRH RLSGMDITLA DYDGRTALHL 540
541 AASEGHLECV KFLLEQCHVP HNPKDRWGNL PVDEAENFGH CHVVEFLRSW AEKADQSSEE 600
601 CKPEAVTSKT QADEEICSTS DLETSPTTSP IPTPVESGSR AGSGRSSPVP SDAGSTASAG 660
661 SSIDDKTKPG L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
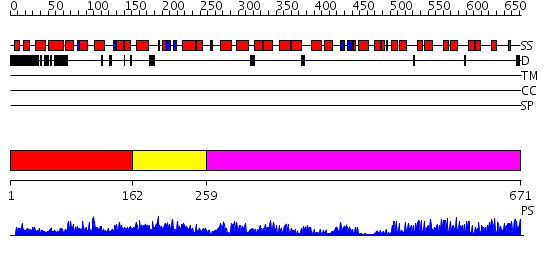
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 1.08 | Solution Structure Of The Calcium-loaded N-Terminal Sensor Domain Of Centrin | |
| 2 | View Details | [162..258] | 43.69897 | No description for 2dfwA was found. | |
| 3 | View Details | [259..671] | 81.69897 | Ankyrin-R |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)