













| General Information: |
|
| Name(s) found: |
FBpp0293023
[FlyBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1918 amino acids |
Gene Ontology: |
|
| Cellular Component: |
filamentous actin
[IDA]
|
| Biological Process: |
bristle morphogenesis
[IMP]
epidermal cell differentiation [IMP] antennal morphogenesis [IMP] imaginal disc-derived wing hair organization [IMP] actin filament bundle assembly [IMP] cuticle pattern formation [IMP] |
| Molecular Function: |
actin binding
[ISS][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASCAPVIVW NGAQFTASSR ELEQLRRDYG LDARMLEATL VPRYKRHKPQ AAARNDHVRV 60
61 TKEQQSDWIL KQDRSTNVSQ VNFHPWAGAH FDLVPWRRTR SALDLTAVED YNHGLVDPKS 120
121 LKGLEKLELR QQRRRLQRAR STHQGLRRLV RVRLIFSIEL RHKRMEVLRK SSLPVRVFEQ 180
181 RFALSENESA AYERILLDAN AGVDQHVGYF SKRQVVTSNR ESNEELVVDF ASETEADEKA 240
241 DEQPEKEEAQ YATIAPAEKG GSAAVPDKES GQVPCVKCRD IAIKLRTIND YALNSNLHLS 300
301 HNTKLNNLHS NKLLHSNQSH SSSINPSTRM HSHQLDSYLG GVGSMMNLGG MGQVGQMYHP 360
361 GSVSGSERPN GGALALHYAA ARGCLDCVQL LVAASVDICY FSTINVKKGL LGRMTTSLTS 420
421 YRRSKQKSKS NNDIHTIIAT EVVTAAILSS SVHSSSTGSE IGDGGQGNTS LIEEAAAQDQ 480
481 SSEEQDRQHQ HAKDPLAQTE MNHHPHSHHQ RKSAVEVVRQ QMDDDGDQDS NDDDETMPMM 540
541 SGVGRKEQNN RRSVVSANTQ MDNDVTPVYL AAQEGHLEVL KFLVLEAGGS LYVRARDGMA 600
601 PIHAASQMGC LDCLKWMVQD QGVDPNLRDG DGATPLHFAA SRGHLSVVRW LLNHGAKLSL 660
661 DKYGKSPIND AAENQQVECL NVLVQHGTSV DYNGKSSSQR HKSQQQLHQQ HQQQQQQQQQ 720
721 QHLSSSCNSN SNSSKQTSRS NTIKSKSSST LSSDVEPFYL HPPALAGGSG GSGMGLGMGM 780
781 GMGMGMSMGL GISRKNSDAL YSQQSRSSSE KLYNGSSSLG GNPGGNSSSG GGGVGGGGGS 840
841 GGALLPNDGL YVNPMRNGGM YNTPSPNGSI SGESFFLHDP QDIIYNRVRD LFVDSDCSSS 900
901 VKQHGKSHAH QLLQSKAGNA MTIQADVHSS SSGAGSGSDE SVSISSSFNS PKQQQQLRSQ 960
961 SFNRHGSSSN ISASGNIKNN NYKAHKGKPS NQNGGGDHDY EDIYLVREES RKQHQQQQQH1020
1021 QLQQQQQLQH QATQQQQQQQ QQQQQLQHKY NVGRSRSRDS GSHSRSASAS STRSTDIVLQ1080
1081 YSNHHLNNKR NMNNNSNMNN SSSNIAQSSS SNNNNNSLLN RNKSHSIIGL HSSKYESSLK1140
1141 DNYSAKNVNL KNQLLNGGIK SDTYESVCPP EDVAERTKQT HKNSMIRNNL ADASSNNNTS1200
1201 GSINNISNIG NMNGGNQSSR NLKRVSSAPP MQNLAVVNGP PPPPLPPPLR APVQQQRNNL1260
1261 SVDQPNSMTG NNGIYKKNVF GPNQGPTNAN GTGGAAPPPV PAPNPVATEA VDSDSGLEVV1320
1321 EEPSLRPSEL VRGNHNRTMS TISANKKAKL LNAGSTNGSS IAASSDDSQS RYGGSVHAAN1380
1381 GSAANGHFYG YSEGNKNQGS NGNGNGNGNG GGGNYHSHPN QPHYATGQPH QYTPSLYGGG1440
1441 SSAEDHYELT QQQQQIYGET HLPSGQRTGG PNLVNKQLVL PFVPPSFPNK SQDGVTHLIK1500
1501 PSEYLKSISI DKRSCPSSAR STDTEDYMQI QIAQQQSHPH QHQHHPHPHP HPHHPHQLHG1560
1561 MGEQPPLPPP PPPLPHGMLP PQPPMGPPMG VAGGQDATMS GKSHKPKKPH GSAPNSQDTA1620
1621 TRKQHQPLSA ISIQDLNSVQ LRRTDTQKVP KPYQMPARSL SMQCLTSSTE TYLKSDLIAE1680
1681 LKISKDIPGI KKMKVEQQMA SRMDSEHYME ITKQFSGNNY VDQIPERDQA GNAIPDWKRQ1740
1741 MMAKKAAERA KKDFEERMAQ EAESRRLSQI PQWKRDLLAR REETENKLKA AIYTPKVEEN1800
1801 NRVADTWRLK NRAMSIDNIN INLAGMEQHY QQIQFQQKQQ QLQLGNKENH VDPKGADQDQ1860
1861 EHGVGIGSGV GNANSGVIGG QELIDPSEGS QGRAEEEEDN IIPWRAQLRK TNSRLSLI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
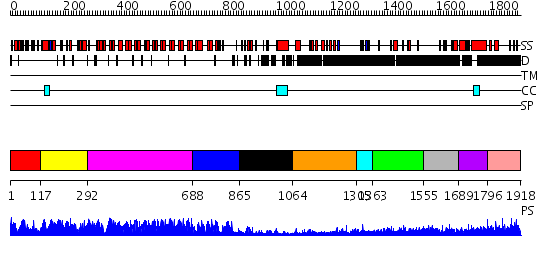
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..291] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [292..687] | 101.0 | Ankyrin-R | |
| 4 | View Details | [688..864] | 34.0 | GA bindinig protein (GABP) beta 1 | |
| 5 | View Details | [865..1063] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1064..1304] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1305..1362] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1363..1554] | 1.204988 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1555..1688] | 4.204996 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1689..1795] | 1.147995 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1796..1918] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)