













| General Information: |
|
| Name(s) found: |
gi|116007158
[NCBI NR]
gi|113193605 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 653 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAICFVCNL PIMSHQVGLV WQGGNGWDDL VREQLEESTI RQRLGGRRDS AAQITTPPST 60
61 SPPPGLQRRR RSSLAQLTDI LREWSGGTKQ QQQHHSQQQQ QQLQQASRSK AQLNRRETLA 120
121 DLARSLPWRT STTTDASTGA GGSGGGGCTT YMAPRKRRES SADSGIRSMR SRRDSNTAEF 180
181 VRHWNRRESG AVEEQPSVSV PVVVECSKSA SRRGSGESYL SSSRRDSNVA GRVTTPPPPG 240
241 KYQMTKKRDS LAAPEFPNFR QEQRPSTSSA GSCSDHSMPL ISTHSSTHHL AHQQVDSACQ 300
301 ALPPPTIITS SVTPPATSPT APKGRRDSQT QCGRVNRRDS KAGVSPERAP RLQRLQRQAT 360
361 AFDEGCLPGG SRRGSQPALS PDPPDDCERR TSRRDSLSPD SASRGGRRDS RTHLSPDRSH 420
421 ERDGSPRRHT LRRQSSSAAR SPRSPDSNSC CSSRDPSPCS RPQTQTTVEQ NQRPAIRRQS 480
481 TTEEILIARG FRRQSTTEEM IRCRNFRRQS SQSDDVCRYR GRRDSSAQII DGTIGTMTVE 540
541 TTSTFFDSST QTEPSPLYDN NHYHEECLRC NSCGLNLTGP NQKRARRFKN QILCDLHFAD 600
601 VALMECSDFM QQLRSFKPQS LGCAVARRKS STTLIFPLPP QACSDLQINR NQF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
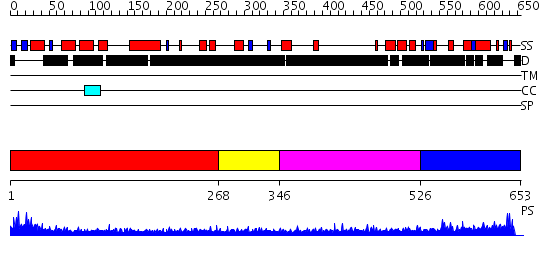
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..267] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [268..345] | 2.150991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [346..525] | 1.146983 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [526..653] | 1.12 | LIM only 4 (Lmo4) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)