













| General Information: |
|
| Name(s) found: |
DYRK3
[HGNC (HUGO)]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 588 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGTARGPGR KDAGPPGAGL PPQQRRLGDG VYDTFMMIDE TKCPPCSNVL CNPSEPPPPR 60
61 RLNMTTEQFT GDHTQHFLDG GEMKVEQLFQ EFGNRKSNTI QSDGISDSEK CSPTVSQGKS 120
121 SDCLNTVKSN SSSKAPKVVP LTPEQALKQY KHHLTAYEKL EIINYPEIYF VGPNAKKRHG 180
181 VIGGPNNGGY DDADGAYIHV PRDHLAYRYE VLKIIGKGSF GQVARVYDHK LRQYVALKMV 240
241 RNEKRFHRQA AEEIRILEHL KKQDKTGSMN VIHMLESFTF RNHVCMAFEL LSIDLYELIK 300
301 KNKFQGFSVQ LVRKFAQSIL QSLDALHKNK IIHCDLKPEN ILLKHHGRSS TKVIDFGSSC 360
361 FEYQKLYTYI QSRFYRAPEI ILGSRYSTPI DIWSFGCILA ELLTGQPLFP GEDEGDQLAC 420
421 MMELLGMPPP KLLEQSKRAK YFINSKGIPR YCSVTTQADG RVVLVGGRSR RGKKRGPPGS 480
481 KDWGTALKGC DDYLFIEFLK RCLHWDPSAR LTPAQALRHP WISKSVPRPL TTIDKVSGKR 540
541 VVNPASAFQG LGSKLPPVVG IANKLKANLM SETNGSIPLC SVLPKLIS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
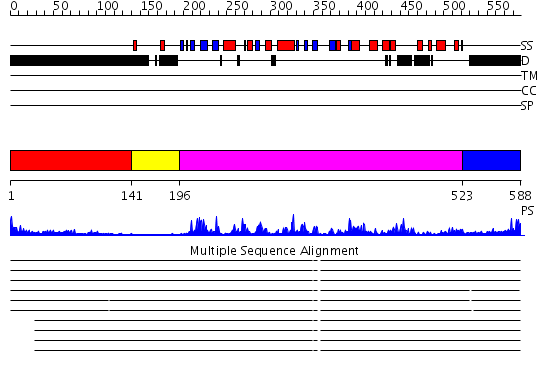
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 0.982 | Riboflavin kinase | |
| 2 | View Details | [141..195] | 47.30103 | No description for 2j0jA was found. | |
| 3 | View Details | [196..522] | 49.09691 | Crystal structure of human CLK1 in complex with 10Z-Hymenialdisine | |
| 4 | View Details | [523..588] | 55.0 | crystal structure of Glycogen synthase kinase 3 in complexed with inhibitor |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)