













| General Information: |
|
| Name(s) found: |
gi|38679290
[NCBI NR]
gi|119630902 [NCBI NR] gi|119630897 [NCBI NR] gi|11641415 [NCBI NR] |
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 601 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEVPQPEPAP GSALSPAGVC GGAQRPGHLP GLLLGSHGLL GSPVRAAASS PVTTLTQTMH 60
61 DLAGLGSRSR LTHLSLSRRA SESSLSSESS ESSDAGLCMD SPSPMDPHMA EQTFEQAIQA 120
121 ASRIIRNEQF AIRRFQSMPV RLLGHSPVLR NITNSQAPDG RRKSEAGSGA ASSSGEDKEN 180
181 VRFWKAGVGA LREEEGACWG GSLACEDPPL PSWLQDGFVF KMPWKPTHPS STHALAEWAS 240
241 RREAFAQRPS SAPDLMCLSP DRKMEVEELS PLALGRFSLT PAEGDTEEDD GFVDILESDL 300
301 KDDDAVPPGM ESLISAPLVK TLEKEEEKDL VMYSKCQRLF RSPSMPCSVI RPILKRLERP 360
361 QDRDTPVQNK RRRSVTPPEE QQEAEEPKAR VLRSKSLCHD EIENLLDSDH RELIGDYSKA 420
421 FLLQTVDGKH QDLKYISPET MVALLTGKFS NIVDKFVIVD CRYPYEYEGG HIKTAVNLPL 480
481 ERDAESFLLK SPIAPCSLDK RVILIFHCEF SSERGPRMCR FIRERDRAVN DYPSLYYPEM 540
541 YILKGGYKEF FPQHPNFCEP QDYRPMNHEA FKDELKTFRL KTRSWAGERS RRELCSRLQD 600
601 Q |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
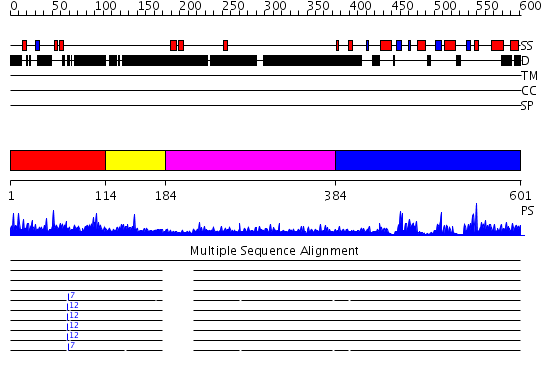
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [114..183] | 1.045993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [184..383] | 1.08499 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [384..601] | 68.0 | CDC25b |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)