













| General Information: |
|
| Name(s) found: |
PRKCA
[HGNC (HUGO)]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 672 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADVFPGNDS TASQDVANRF ARKGALRQKN VHEVKDHKFI ARFFKQPTFC SHCTDFIWGF 60
61 GKQGFQCQVC CFVVHKRCHE FVTFSCPGAD KGPDTDDPRS KHKFKIHTYG SPTFCDHCGS 120
121 LLYGLIHQGM KCDTCDMNVH KQCVINVPSL CGMDHTEKRG RIYLKAEVAD EKLHVTVRDA 180
181 KNLIPMDPNG LSDPYVKLKL IPDPKNESKQ KTKTIRSTLN PQWNESFTFK LKPSDKDRRL 240
241 SVEIWDWDRT TRNDFMGSLS FGVSELMKMP ASGWYKLLNQ EEGEYYNVPI PEGDEEGNME 300
301 LRQKFEKAKL GPAGNKVISP SEDRKQPSNN LDRVKLTDFN FLMVLGKGSF GKVMLADRKG 360
361 TEELYAIKIL KKDVVIQDDD VECTMVEKRV LALLDKPPFL TQLHSCFQTV DRLYFVMEYV 420
421 NGGDLMYHIQ QVGKFKEPQA VFYAAEISIG LFFLHKRGII YRDLKLDNVM LDSEGHIKIA 480
481 DFGMCKEHMM DGVTTRTFCG TPDYIAPEII AYQPYGKSVD WWAYGVLLYE MLAGQPPFDG 540
541 EDEDELFQSI MEHNVSYPKS LSKEAVSICK GLMTKHPAKR LGCGPEGERD VREHAFFRRI 600
601 DWEKLENREI QPPFKPKVCG KGAENFDKFF TRGQPVLTPP DQLVIANIDQ SDFEGFSYVN 660
661 PQFVHPILQS AV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
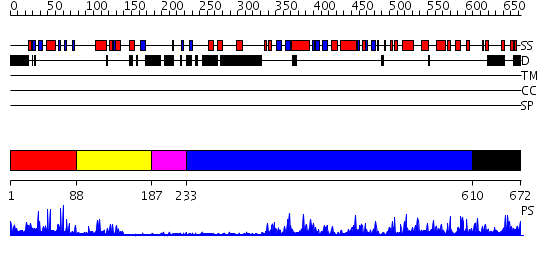
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 9.69897 | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | |
| 2 | View Details | [88..186] | 3.5 | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | |
| 3 | View Details | [187..232] | 6.0 | No description for PF00168.21 was found. No confident structure predictions are available. | |
| 4 | View Details | [233..609] | 41.0 | Carboxyl-terminal src kinase (csk) | |
| 5 | View Details | [610..672] | 96.045757 | No description for 2i0eA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)