













| General Information: |
|
| Name(s) found: |
TUBGCP2
[HGNC (HUGO)]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 902 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEFRIHHDV NELLSLLRVH GGDGAEVYID LLQKNRTPYV TTTVSAHSAK VKIAEFSRTP 60
61 EDFLKKYDEL KSKNTRNLDP LVYLLSKLTE DKETLQYLQQ NAKERAELAA AAVGSSTTSI 120
121 NVPAAASKIS MQELEELRKQ LGSVATGSTL QQSLELKRKM LRDKQNKKNS GQHLPIFPAW 180
181 VYERPALIGD FLIGAGISTD TALPIGTLPL ASQESAVVED LLYVLVGVDG RYVSAQPLAG 240
241 RQSRTFLVDP NLDLSIRELV HRILPVAASY SAVTRFIEEK SSFEYGQVNH ALAAAMRTLV 300
301 KEHLILVSQL EQLHRQGLLS LQKLWFYIQP AMRTMDILAS LATSVDKGEC LGGSTLSLLH 360
361 DRSFSYTGDS QAQELCLYLT KAASAPYFEV LEKWIYRGII HDPYSEFMVE EHELRKERIQ 420
421 EDYNDKYWDQ RYTIVQQQIP SFLQKMADKI LSTGKYLNVV RECGHDVTCP VAKEIIYTLK 480
481 ERAYVEQIEK AFNYASKVLL DFLMEEKELV AHLRSIKRYF LMDQGDFFVH FMDLAEEELR 540
541 KPVEDITPPR LEALLELALR MSTANTDPFK DDLKIDLMPH DLITQLLRVL AIETKQEKAM 600
601 AHADPTELAL SGLEAFSFDY IVKWPLSLII NRKALTRYQM LFRHMFYCKH VERQLCSVWI 660
661 SNKTAKQHSL HSAQWFAGAF TLRQRMLNFV QNIQYYMMFE VMEPTWHILE KNLKSASNID 720
721 DVLGHHTGFL DTCLKDCMLT NPELLKVFSK LMSVCVMFTN CMQKFTQSMK LDGELGGQTL 780
781 EHSTVLGLPA GAEERARKEL ARKHLAEHAD TVQLVSGFEA TINKFDKNFS AHLLDLLARL 840
841 SIYSTSDCEH GMASVISRLD FNGFYTERLE RLSAERSQKA TPQVPVLRGP PAPAPRVAVT 900
901 AQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
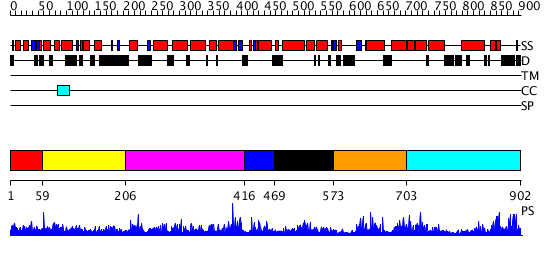
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..58] | 1.012982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [59..205] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [206..415] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [416..468] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [469..572] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [573..702] | 1.098991 | View MSA. Confident ab initio structure predictions are available. | |
| 7 | View Details | [703..902] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)