













| General Information: |
|
| Name(s) found: |
gi|222624660
[NCBI NR]
gi|215768510 [NCBI NR] gi|115452221 [NCBI NR] gi|113548182 [NCBI NR] gi|108707460 [NCBI NR] |
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 726 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEEAKPNPN ADAAPAAAEA AASSPPPLSS SESAEGKDKE EEKEEEEEEE AGDLVEKLVE 60
61 LVEEIAAISD FRNAYRRQFC NLSRRIRLLA PMLEEAKEGP RPLPDVSYSA LRRLREALAD 120
121 SRELLRLGVS GSKISLVLER EKIMKSFQDI TARLEQALGL ISFDELDISD EVREQVELVH 180
181 AQFKRAKERS DPSDDDLFND LVSVYNSSTS ANVDPDILQR LSDKLQLATI SDLNQESLIL 240
241 HEMASGGDPG AVVEKMSMLL KRIKDFVQSR DPEMGTPVNT TELSGKDNMA SLIVPDDFRC 300
301 PISLDLMKDP VIVATGQTYE RGCIERWLEA GHDTCPKTQQ KLPNKSLTPN YVLRSLIAQW 360
361 CEANGMEPPK RAAQHHNAPA SCTAAEHSNV VELLQKLLSQ NLEDQREAAG MLRQLAKRSP 420
421 ENRACIGDAG AIPILVSLLS ITDVSTQEHV VTALLNLSIY EENKARIITS GAVPGVVHVL 480
481 KRGSMEAREN SAATLFSLSL VDENKITIGA SGAIPALVLL LSNGSQRGKR DAATALFNLC 540
541 IYQGNKGKAV RAGLIPVLLG LVTETESGMM DEALAILAIL SSHPEGKTAI SSANAIPMLV 600
601 GVIRNGSARN KENAAAVLVH LCNGEQQQQH LAEAQEQGIV TLLEELAKSG TDRGKRKAIQ 660
661 LLERMNRFLM QQSQAQAQAE AMAQAHAHAQ SQAQVQALNE AQSQVEMQVE QLLLPTTSHL 720
721 SDRRDG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
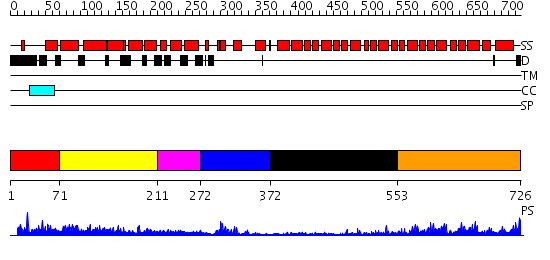
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [71..210] | 18.0 | Crystal structure of the CHIP U-box E3 ubiquitin ligase | |
| 3 | View Details | [211..271] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [272..371] | 8.69897 | NMR solution structure of the U box domain from AtPUB14, an armadillo repeat containing protein from Arabidopsis thaliana | |
| 5 | View Details | [372..552] | 3.59 | Crystal structure of the HspBP1 core domain | |
| 6 | View Details | [553..726] | 4.69897 | AP1 clathrin adaptor core |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 |
|
||||||
| 5 |
|
||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)