













| General Information: |
|
| Name(s) found: |
TRMT2B
[HGNC (HUGO)]
TRIM2 [HGNC (HUGO)] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 744 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASEGTNIPS PVVRQIDKQF LICSICLERY KNPKVLPCLH TFCERCLQNY IPAHSLTLSC 60
61 PVCRQTSILP EKGVAALQNN FFITNLMDVL QRTPGSNAEE SSILETVTAV AAGKPLSCPN 120
121 HDGNVMEFYC QSCETAMCRE CTEGEHAEHP TVPLKDVVEQ HKASLQVQLD AVNKRLPEID 180
181 SALQFISEII HQLTNQKASI VDDIHSTFDE LQKTLNVRKS VLLMELEVNY GLKHKVLQSQ 240
241 LDTLLQGQES IKSCSNFTAQ ALNHGTETEV LLVKKQMSEK LNELADQDFP LHPRENDQLD 300
301 FIVETEGLKK SIHNLGTILT TNAVASETVA TGEGLRQTII GQPMSVTITT KDKDGELCKT 360
361 GNAYLTAELS TPDGSVADGE ILDNKNGTYE FLYTVQKEGD FTLSLRLYDQ HIRGSPFKLK 420
421 VIRSADVSPT TEGVKRRVKS PGSGHVKQKA VKRPASMYST GKRKENPIED DLIFRVGTKG 480
481 RNKGEFTNLQ GVAASTNGKI LIADSNNQCV QIFSNDGQFK SRFGIRGRSP GQLQRPTGVA 540
541 VHPSGDIIIA DYDNKWVSIF SSDGKFKTKI GSGKLMGPKG VSVDRNGHII VVDNKACCVF 600
601 IFQPNGKIVT RFGSRGNGDR QFAGPHFAAV NSNNEIIITD FHNHSVKVFN QEGEFMLKFG 660
661 SNGEGNGQFN APTGVAVDSN GNIIVADWGN SRIQVFDGSG SFLSYINTSA DPLYGPQGLA 720
721 LTSDGHVVVA DSGNHCFKVY RYLQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
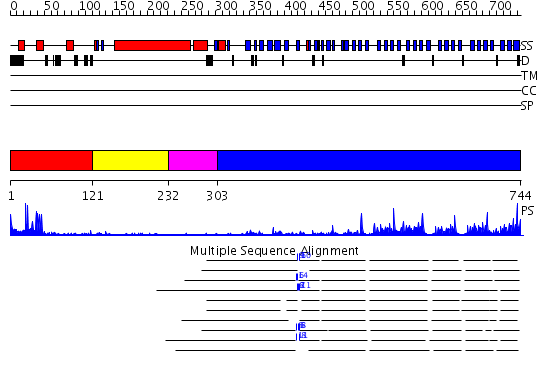
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 4.045757 | No description for 2cklB was found. | |
| 2 | View Details | [121..231] | 2.54 | No description for 2djaA was found. | |
| 3 | View Details | [232..302] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [303..744] | 34.09691 | Actin interacting protein 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)