













| General Information: |
|
| Name(s) found: |
DPP3
[HGNC (HUGO)]
|
| Description(s) found:
Found 37 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 737 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADTQYILPN DIGVSSLDCR EAFRLLSPTE RLYAYHLSRA AWYGGLAVLL QTSPEAPYIY 60
61 ALLSRLFRAQ DPDQLRQHAL AEGLTEEEYQ AFLVYAAGVY SNMGNYKSFG DTKFVPNLPK 120
121 EKLERVILGS EAAQQHPEEV RGLWQTCGEL MFSLEPRLRH LGLGKEGITT YFSGNCTMED 180
181 AKLAQDFLDS QNLSAYNTRL FKEVDGEGKP YYEVRLASVL GSEPSLDSEV TSKLKSYEFR 240
241 GSPFQVTRGD YAPILQKVVE QLEKAKAYAA NSHQGQMLAQ YIESFTQGSI EAHKRGSRFW 300
301 IQDKGPIVES YIGFIESYRD PFGSRGEFEG FVAVVNKAMS AKFERLVASA EQLLKELPWP 360
361 PTFEKDKFLT PDFTSLDVLT FAGSGIPAGI NIPNYDDLRQ TEGFKNVSLG NVLAVAYATQ 420
421 REKLTFLEED DKDLYILWKG PSFDVQVGLH ELLGHGSGKL FVQDEKGAFN FDQETVINPE 480
481 TGEQIQSWYR SGETWDSKFS TIASSYEECR AESVGLYLCL HPQVLEIFGF EGADAEDVIY 540
541 VNWLNMVRAG LLALEFYTPE AFNWRQAHMQ ARFVILRVLL EAGEGLVTIT PTTGSDGRPD 600
601 ARVRLDRSKI RSVGKPALER FLRRLQVLKS TGDVAGGRAL YEGYATVTDA PPECFLTLRD 660
661 TVLLRKESRK LIVQPNTRLE GSDVQLLEYE ASAAGLIRSF SERFPEDGPE LEEILTQLAT 720
721 ADARFWKGPS EAPSGQA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
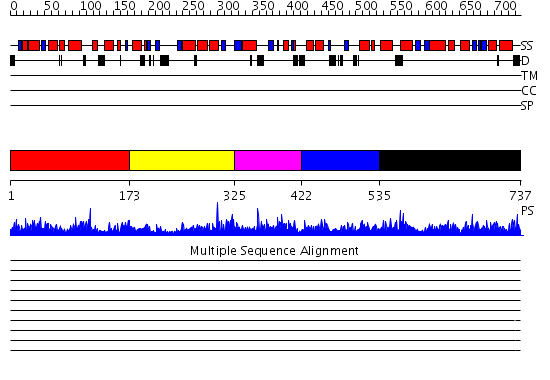
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 2.041942 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [173..324] | 1.055971 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [325..421] | 1.058947 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [422..534] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [535..737] | 1.058975 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)