













| General Information: |
|
| Name(s) found: |
C2orf44
[HGNC (HUGO)]
|
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 721 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELGKGKLLR TGLNALHQAV HPIHGLAWTD GNQVVLTDLR LHSGEVKFGD SKVIGQFECV 60
61 CGLSWAPPVA DDTPVLLAVQ HEKHVTVWQL CPSPMESSKW LTSQTCEIRG SLPILPQGCV 120
121 WHPKCAILTV LTAQDVSIFP NVHSDDSQVK ADINTQGRIH CACWTQDGLR LVVAVGSSLH 180
181 SYIWDSAQKT LHRCSSCLVF DVDSHVCSIT ATVDSQVAIA TELPLDKICG LNASETFNIP 240
241 PNSKDMTPYA LPVIGEVRSM DKEATDSETN SEVSVSSSYL EPLDLTHIHF NQHKSEGNSL 300
301 ICLRKKDYLT GTGQDSSHLV LVTFKKAVTM TRKVTIPGIL VPDLIAFNLK AHVVAVASNT 360
361 CNIILIYSVI PSSVPNIQQI RLENTERPKG ICFLTDQLLL ILVGKQKLTD TTFLPSSKSD 420
421 QYAISLIVRE IMLEEEPSIT SGESQTTYST FSAPLNKANR KKLIESLSPD FCHQNKGLLL 480
481 TVNTSSQNGR PGRTLIKEIQ SPLSSICDGS IALDAEPVTQ PASLPRHSST PDHTSTLEPP 540
541 RLPQRKNLQS EKETYQLSKE VEILSRNLVE MQRCLSELTN RLHNGKKSSS VYPLSQDLPY 600
601 VHIIYQKPYY LGPVVEKRAV LLCDGKLRLS TVQQTFGLSL IEMLHDSHWI LLSADSEGFI 660
661 PLTFTATQEI IIRDGSLSRS DVFRDSFSHS PGAVSSLKVF TGLAAPSLDT TGCCNHVDGM 720
721 A |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
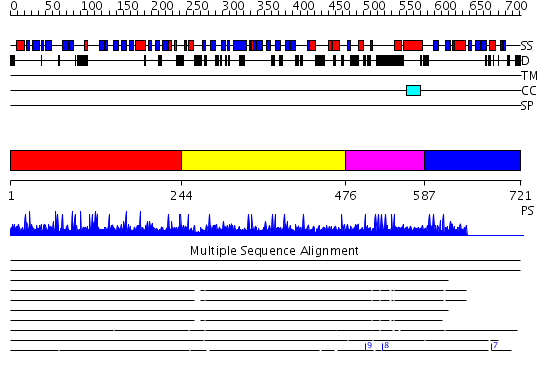
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..243] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [244..475] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [476..586] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [587..721] | 1.010995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)