













| General Information: |
|
| Name(s) found: |
COPB2
[HGNC (HUGO)]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 906 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLRLDIKRK LTARSDRVKS VDLHPTEPWM LASLYNGSVC VWNHETQTLV KTFEVCDLPV 60
61 RAAKFVARKN WVVTGADDMQ IRVFNYNTLE RVHMFEAHSD YIRCIAVHPT QPFILTSSDD 120
121 MLIKLWDWDK KWSCSQVFEG HTHYVMQIVI NPKDNNQFAS ASLDRTIKVW QLGSSSPNFT 180
181 LEGHEKGVNC IDYYSGGDKP YLISGADDRL VKIWDYQNKT CVQTLEGHAQ NVSCASFHPE 240
241 LPIIITGSED GTVRIWHSST YRLESTLNYG MERVWCVASL RGSNNVALGY DEGSIIVKLG 300
301 REEPAMSMDA NGKIIWAKHS EVQQANLKAM GDAEIKDGER LPLAVKDMGS CEIYPQTIQH 360
361 NPNGRFVVVC GDGEYIIYTA MALRNKSFGS AQEFAWAHDS SEYAIRESNS IVKIFKNFKE 420
421 KKSFKPDFGA ESIYGGFLLG VRSVNGLAFY DWDNTELIRR IEIQPKHIFW SDSGELVCIA 480
481 TEESFFILKY LSEKVLAAQE THEGVTEDGI EDAFEVLGEI QEIVKTGLWV GDCFIYTSSV 540
541 NRLNYYVGGE IVTIAHLDRT MYLLGYIPKD NRLYLGDKEL NIISYSLLVS VLEYQTAVMR 600
601 RDFSMADKVL PTIPKEQRTR VAHFLEKQGF KQQALTVSTD PEHRFELALQ LGELKIAYQL 660
661 AVEAESEQKW KQLAELAISK CQFGLAQECL HHAQDYGGLL LLATASGNAN MVNKLAEGAE 720
721 RDGKNNVAFM SYFLQGKVDA CLELLIRTGR LPEAAFLART YLPSQVSRVV KLWRENLSKV 780
781 NQKAAESLAD PTEYENLFPG LKEAFVVEEW VKETHADLWP AKQYPLVTPN EERNVMEEGK 840
841 DFQPSRSTAQ QELDGKPASP TPVIVASHTA NKEEKSLLEL EVDLDNLELE DIDTTDINLD 900
901 EDILDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
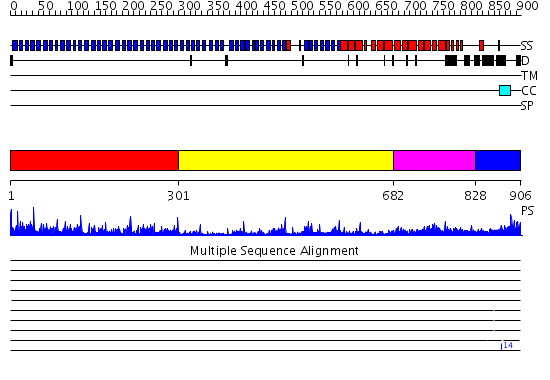
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..300] | 71.045757 | No description for 2gnqA was found. | |
| 2 | View Details | [301..681] | 4.09 | Cdc4 F-box and linker domains; Cdc4 propeller domain | |
| 3 | View Details | [682..827] | 4.221849 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 4 | View Details | [828..906] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)