













| General Information: |
|
| Name(s) found: |
EPC1
[HGNC (HUGO)]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 836 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKLSFRARA LDASKPLPVF RCEDLPDLHE YASINRAVPQ MPTGMEKEEE SEHHLQRAIS 60
61 AQQVYGEKRD NMVIPVPEAE SNIAYYESIY PGEFKMPKQL IHIQPFSLDA EQPDYDLDSE 120
121 DEVFVNKLKK KMDICPLQFE EMIDRLEKGS GQQPVSLQEA KLLLKEDDEL IREVYEYWIK 180
181 KRKNCRGPSL IPSVKQEKRD GSSTNDPYVA FRRRTEKMQT RKNRKNDEAS YEKMLKLRRD 240
241 LSRAVTILEM IKRREKSKRE LLHLTLEIME KRYNLGDYNG EIMSEVMAQR QPMKPTYAIP 300
301 IIPITNSSQF KHQEAMDVKE FKVNKQDKAD LIRPKRKYEK KPKVLPSSAA ATPQQTSPAA 360
361 LPVFNAKDLN QYDFPSSDEE PLSQVLSGSS EAEEDNDPDG PFAFRRKAGC QYYAPHLDQT 420
421 GNWPWTSPKD GGLGDVRYRY CLTTLTVPQR CIGFARRRVG RGGRVLLDRA HSDYDSVFHH 480
481 LDLEMLSSPQ HSPVNQFANT SETNTSDKSF SKDLSQILVN IKSCRWRHFR PRTPSLHDSD 540
541 NDELSCRKLY RSINRTGTAQ PGTQTCSTST QSKSSSGSAH FAFTAEQYQQ HQQQLALMQK 600
601 QQLAQIQQQQ ANSNSSTNTS QNLASNQQKS GFRLNIQGLE RTLQGFVSKT LDSASAQFAA 660
661 SALVTSEQLM GFKMKDDVVL GIGVNGVLPA SGVYKGLHLS STTPTALVHT SPSTAGSALL 720
721 QPSNITQTSS SHSALSHQVT AANSATTQVL IGNNIRLTVP SSVATVNSIA PINARHIPRT 780
781 LSAVPSSALK LAAAANCQVS KVPSSSSVDS VPRENHESEK PALNNIADNT VAMEVT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
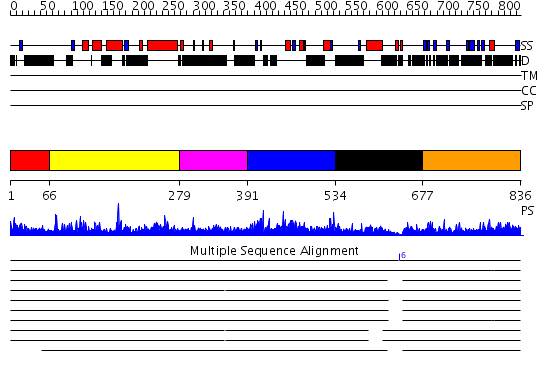
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..65] | 4.051995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [66..278] | 12.107984 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [279..390] | 21.39794 | No description for 2d9eA was found. | |
| 4 | View Details | [391..533] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [534..676] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [677..836] | 2.018949 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)