













| General Information: |
|
| Name(s) found: |
dnj-27 /
CE40162
[WormBase]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 318 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IEA |
| Biological Process: |
response to heat
[IEA glycerol ether metabolic process [IEA cell redox homeostasis [IEA protein folding [IEA |
| Molecular Function: |
heat shock protein binding
[IEA ATP binding [IEA protein disulfide oxidoreductase activity [IEA electron carrier activity [IEA unfolded protein binding [IEA isomerase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLLGEYRRF HTATSEDSML HTVAIGSLDC VKYKDLCQQA GVQSYPTSIV YTPDGKTHKM 60
61 VGYHNVDYIL EFLDNSLNPS VMEMSPEQFE ELVMNRKDEE TWLVDFFAPW CGPCQQLAPE 120
121 LQKAARQIAA FDENAHVASI DCQKYAQFCT NTQINSYPTV RMYPAKKTKQ PRRSPFYDYP 180
181 NHMWRNSDSI QRWVYNFLPT EVVSLGNDFH TTVLDSSEPW IVDFFAPWCG HCIQFAPIYD 240
241 QIAKELAGKV NFAKIDCDQW PGVCQGAQVR AYPTIRLYTG KTGWSRQGDQ GIGIGTQHKE 300
301 QFIQIVRQQL KLDEHDEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
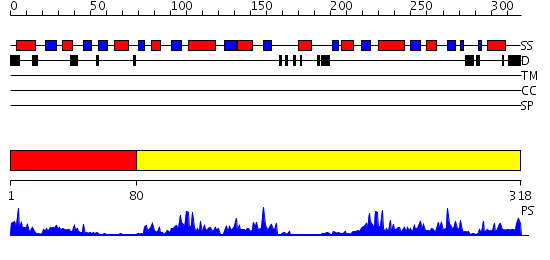
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | 8.522879 | Protein disulfide isomerase, PDI | |
| 2 | View Details | [80..318] | 22.0 | Crystal Structure of Yeast Protein Disulfide Isomerase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)