













| General Information: |
|
| Name(s) found: |
gi|6330197
[NCBI NR]
|
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 838 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 LTSSMEDPAA PGTGGPPANG NGNGGGKGKQ AAPKGREAFR SQRRESEGSV DCPTLEFEYG 60
61 DADGHAAELS ELYSYTENLE FTNNRRCFEE DFKTQVQGKE WLELEEDAQK AYIMGLLDRL 120
121 EVVSRERRLK VARAVLYLAQ GTFGECDSEV DVLHWSRYNC FLLYQMGTFS TFLELLHMEI 180
181 DNSQACSSAL RKPAVSIADS TELRVLLSVM YLMVENIRLE RETDPCGWRT ARETFRTELS 240
241 FSMHNEEPFA LLLFSMVTKF CSGLAPHFPI KKVLLLLWKV VMFTLGGFEH LQTLKVQKRA 300
301 ELGLPPLAED SIQVVKSMRA ASPPSYTLDL GESQLAPPPS KLRGRRGSRR QLLTKQDSLD 360
361 IYNERDLFKT EEPATEEEEE SAGDGERTLD GELDLLEQDP LVPPPPSQAP LSAERVAFPK 420
421 GLPWAPKVRQ KDIEHFLEMS RNKFIGFTLG QDTDTLVGLP RPIHESVKTL KQHKYISIAD 480
481 VQIKNEEELE KCPMSLGEEV VPETPCEILY QGMLYSLPQY MIALLKILLA AAPTSKAKTD 540
541 SINILADVLP EEMPITVLQS MKLGIDVNRH KEIIVKSIST LLLLLLKHFK LNHIYQFEYV 600
601 SQHLVFANCI PLILKFFNQN ILSYITAKNS ISVLDYPCCT IQDLPELTTE SLEAGDNSQF 660
661 CWRNLFSCIN LLRLLNKLTK WKHSRTMMLV VFKSAPILKR ALKVKQAMLQ LYVLKLLKLQ 720
721 TKYLGRQWRK SNMKTMSAIY QKVRHRMNDD WAYGNDIDAR PWDFQAEECT LRANIEAFNS 780
781 RRYDRPQDSE FSPVDNCLQS VLGQRLDLPE DFHYSYELWL EREVFSQPIC WEELLQNH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
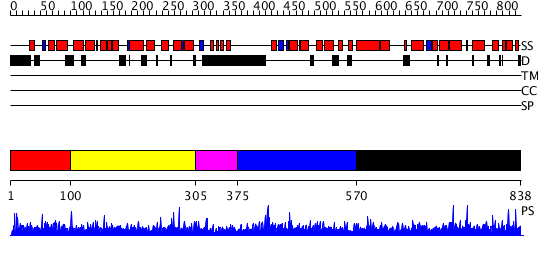
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..304] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [305..374] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [375..569] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [570..838] | 1.016941 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)