













| General Information: |
|
| Name(s) found: |
C11orf2
[HGNC (HUGO)]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 782 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAAAAAGPS PGSGPGDSPE GPEGEAPERR RKAHGMLKLY YGLSEGEAAG RPAGPDPLDP 60
61 TDLNGAHFDP EVYLDKLRRE CPLAQLMDSE TDMVRQIRAL DSDMQTLVYE NYNKFISATD 120
121 TIRKMKNDFR KMEDEMDRLA TNMAVITDFS ARISATLQDR HERITKLAGV HALLRKLQFL 180
181 FELPSRLTKC VELGAYGQAV RYQGRAQAVL QQYQHLPSFR AIQDDCQVIT ARLAQQLRQR 240
241 FREGGSGAPE QAECVELLLA LGEPAEELCE EFLAHARGRL EKELRNLEAE LGPSPPAPDV 300
301 LEFTDHGGSG FVGGLCQVAA AYQELFAAQG PAGAEKLAAF ARQLGSRYFA LVERRLAQEQ 360
361 GGGDNSLLVR ALDRFHRRLR APGALLAAAG LADAATEIVE RVARERLGHH LQGLRAAFLG 420
421 CLTDVRQALA APRVAGKEGP GLAELLANVA SSILSHIKAS LAAVHLFTAK EVSFSNKPYF 480
481 RGEFCSQGVR EGLIVGFVHS MCQTAQSFCD SPGEKGGATP PALLLLLSRL CLDYETATIS 540
541 YILTLTDEQF LVQDQFPVTP VSTLCAEARE TARRLLTHYV KVQGLVISQM LRKSVETRDW 600
601 LSTLEPRNVR AVMKRVVEDT TAIDVQVGLL YEEGVRKAQS SDSSKRTFSV YSSSRQQGRY 660
661 APSYTPSAPM DTNLLSNIQK LFSERIDVFS PVEFNKVSVL TGIIKISLKT LLECVRLRTF 720
721 GRFGLQQVQV DCHFLQLYLW RFVADEELVH LLLDEVVASA ALRCPDPVPM EPSVVEVICE 780
781 RG |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
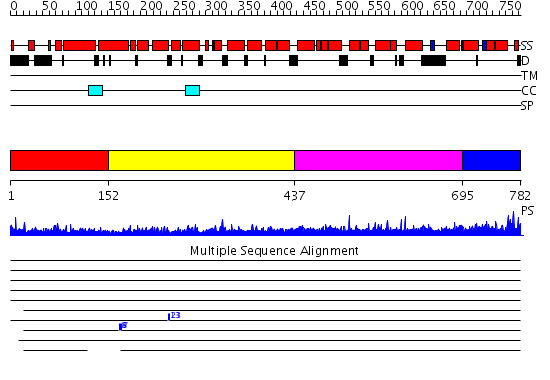
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 35.431798 | No description for PF08700.2 was found. No confident structure predictions are available. | |
| 2 | View Details | [152..436] | 3.69897 | Crystal Structure of the Exo84p C-terminal Domains | |
| 3 | View Details | [437..694] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [695..782] | 2.020974 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)