













| General Information: |
|
| Name(s) found: |
COG2
[HGNC (HUGO)]
|
| Description(s) found:
Found 39 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 738 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKSRMNLPK GPDTLCFDKD EFMKEDFDVD HFVSDCRKRV QLEELRDDLE LYYKLLKTAM 60
61 VELINKDYAD FVNLSTNLVG MDKALNQLSV PLGQLREEVL SLRSSVSEGI RAVDERMSKQ 120
121 EDIRKKKMCV LRLIQVIRSV EKIEKILNSQ SSKETSALEA SSPLLTGQIL ERIATEFNQL 180
181 QFHAVQSKGM PLLDKVRPRI AGITAMLQQS LEGLLLEGLQ TSDVDIIRHC LRTYATIDKT 240
241 RDAEALVGQV LVKPYIDEVI IEQFVESHPN GLQVMYNKLL EFVPHHCRLL REVTGGAISS 300
301 EKGNTVPGYD FLVNSVWPQI VQGLEEKLPS LFNPGNPDAF HEKYTISMDF VRRLERQCGS 360
361 QASVKRLRAH PAYHSFNKKW NLPVYFQIRF REIAGSLEAA LTDVLEDAPA ESPYCLLASH 420
421 RTWSSLRRCW SDEMFLPLLV HRLWRLTLQI LARYSVFVNE LSLRPISNES PKEIKKPLVT 480
481 GSKEPSITQG NTEDQGSGPS ETKPVVSISR TQLVYVVADL DKLQEQLPEL LEIIKPKLEM 540
541 IGFKNFSSIS AALEDSQSSF SACVPSLSSK IIQDLSDSCF GFLKSALEVP RLYRRTNKEV 600
601 PTTASSYVDS ALKPLFQLQS GHKDKLKQAI IQQWLEGTLS ESTHKYYETV SDVLNSVKKM 660
661 EESLKRLKQA RKTTPANPVG PSGGMSDDDK IRLQLALDVE YLGEQIQKLG LQASDIKSFS 720
721 ALAELVAAAK DQATAEQP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
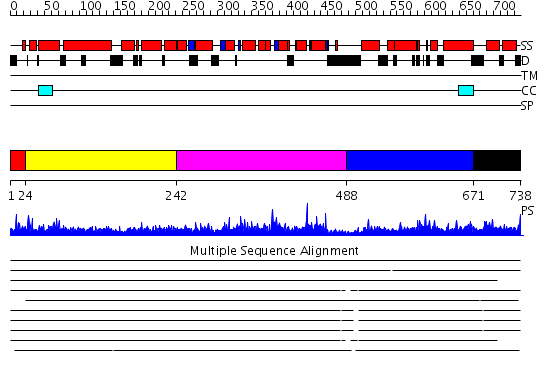
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..23] | 3.107996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [24..241] | 28.045757 | No description for 2jqqA was found. | |
| 3 | View Details | [242..487] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [488..670] | 1.025994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [671..738] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)