













| General Information: |
|
| Name(s) found: |
AMPD2
[HGNC (HUGO)]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 879 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRNRGQGLFR LRSRCFLHQS LPLGAGRRKG LDVAEPGPSR CRSDSPAVAA VVPAMASYPS 60
61 GSGKPKAKYP FKKRASLQAS TAAPEARGGL GAPPLQSARS LPGPAPCLKH FPLDLRTSMD 120
121 GKCKEIAEEL FTRSLAESEL RSAPYEFPEE SPIEQLEERR QRLERQISQD VKLEPDILLR 180
181 AKQDFLKTDS DSDLQLYKEQ GEGQGDRSLR ERDVLEREFQ RVTISGEEKC GVPFTDLLDA 240
241 AKSVVRALFI REKYMALSLQ SFCPTTRRYL QQLAEKPLET RTYEQGPDTP VSADAPVHPP 300
301 ALEQHPYEHC EPSTMPGDLG LGLRMVRGVV HVYTRREPDE HCSEVELPYP DLQEFVADVN 360
361 VLMALIINGP IKSFCYRRLQ YLSSKFQMHV LLNEMKELAA QKKVPHRDFY NIRKVDTHIH 420
421 ASSCMNQKHL LRFIKRAMKR HLEEIVHVEQ GREQTLREVF ESMNLTAYDL SVDTLDVHAD 480
481 RNTFHRFDKF NAKYNPIGES VLREIFIKTD NRVSGKYFAH IIKEVMSDLE ESKYQNAELR 540
541 LSIYGRSRDE WDKLARWAVM HRVHSPNVRW LVQVPRLFDV YRTKGQLANF QEMLENIFLP 600
601 LFEATVHPAS HPELHLFLEH VDGFDSVDDE SKPENHVFNL ESPLPEAWVE EDNPPYAYYL 660
661 YYTFANMAML NHLRRQRGFH TFVLRPHCGE AGPIHHLVSA FMLAENISHG LLLRKAPVLQ 720
721 YLYYLAQIGI AMSPLSNNSL FLSYHRNPLP EYLSRGLMVS LSTDDPLQFH FTKEPLMEEY 780
781 SIATQVWKLS SCDMCELARN SVLMSGFSHK VKSHWLGPNY TKEGPEGNDI RRTNVPDIRV 840
841 GYRYETLCQE LALITQAVQS EMLETIPEEA GITMSPGPQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
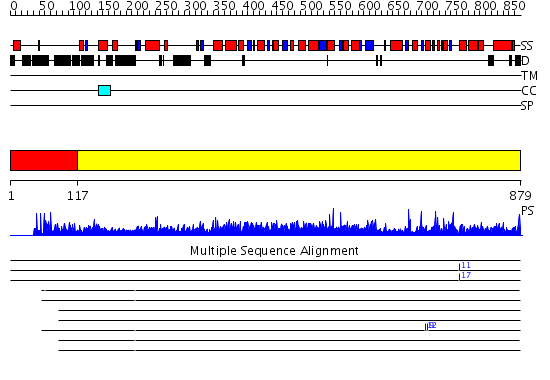
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..879] | 1000.0 | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)