













| General Information: |
|
| Name(s) found: |
RRP46 /
YGR095C
[SGD]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 256 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasmic exosome (RNase complex)
[IDA nuclear exosome (RNase complex) [IDA nuclear outer membrane [TAS |
| Biological Process: |
mRNA catabolic process
[IPI |
| Molecular Function: |
3'-5'-exoribonuclease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNQLRCIKYL CIYGNNKEPN TKNRLDSAEK KKKMSVQAEI GILDHVDGSS EFVSQDTKVI 60
61 CSVTGPIEPK ARQELPTQLA LEIIVRPAKG VATTREKVLE DKLRAVLTPL ITRHCYPRQL 120
121 CQITCQILES GEDEAEFSLR ELSCCINAAF LALVDAGIAL NSMCASIPIA IIKDTSDIIV 180
181 DPTAEQLKIS LSVHTLALEF VNGGKVVKNV LLLDSNGDFN EDQLFSLLEL GEQKCQELVT 240
241 NIRRIIQDNI SPRLVV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
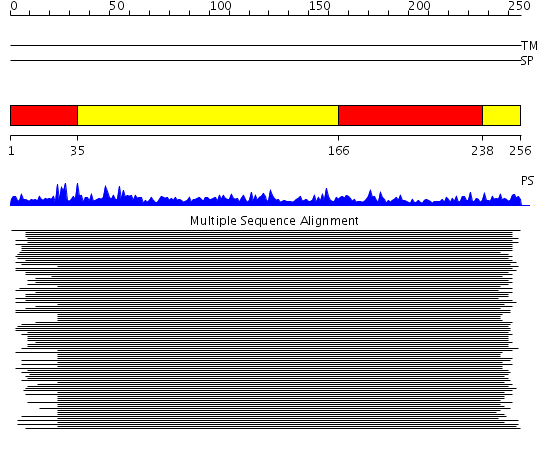
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..34] [166..237] |
49.69897 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [35..165] [238..256] |
49.69897 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)