













| General Information: |
|
| Name(s) found: |
NUP60_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 736 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSGPIRTLH KGKAARNRTP YDRIAASKDG NHSNGPQTPS KSIFQRAKEW LTPSSWKKAI 60
61 SIFSSPVVNK HEDSFDSKTD EEYLQNVSTT TEDVSMLINT PVTEKYEQEH RDTSAQATPS 120
121 IVEQSPNQML ANFFSKKGKT PLNEIEKEGI ISILNKSASP SSSVISPAAS LNRFQTPRAA 180
181 AISKRESGVS SEPRARTSSL TPGNTPNSAK QWSAFRSTFS PLREQDQLST ISPNSLLPAQ 240
241 RLSYYGPTLS TPYNRRLRHK RHSTTPISLS NSIAPSLSFQ PKKARYESAN VSFNDTSFTN 300
301 VPTSSPLHQS TTANHPEKTP SRAAASLLSI LDSKEKNTPS ITAKAGSPQS APSKASYISP 360
361 YARPGITTSR RRHDQIRPSS EKSEPEKKEP SAFETLEKSS NVQTYKPSLM PEFLEKASTH 420
421 GSFAKQKEGE QTSLSEKTAL SEPENKTPVF SFKAPSATTD KPSPPVSSIF SFNAPSAAST 480
481 KPSPAVSSTF SFNAPTTTPS ATSFSIINKE KPARSPNETI DVDLEEEGSG ISAEVEVANE 540
541 GEDLQKNATE VKASTSEKPV FRFEAVTDEK NSEVSSSNQA SSSTMISQPN TGFSFGSFNK 600
601 PAGQEEKPQQ RSLFSASFTT QKPELPAAKI EPEVQMTNVA IDQRSFEQAE KSPISVSEST 660
661 SLVEVEKPSA EGTNEHKQDA TMTLEKTDKQ GSLEEEPFPK FSFTVLPKEN GENLSTMEST 720
721 QELPKFSFSV LKEEKN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
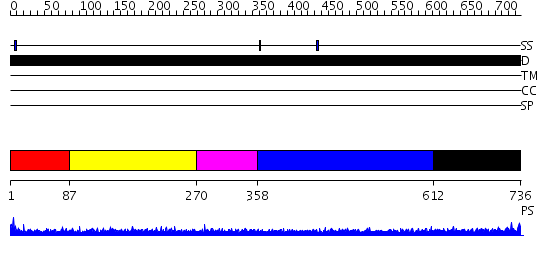
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..269] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [270..357] | 4.190995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [358..611] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [612..736] | 3.172992 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)