













| General Information: |
|
| Name(s) found: |
ORR22_ORYSJ
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 696 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLGALRMEE RKGLMGRERD QFPVGMRVLA VDDDPVCLKV LETLLRRCQY HVTSTNQAIT 60
61 ALKLLRENRD MFDLVISDVH MPDMDGFKLL ELVGLEMDLP VIMLSVNGET KTVMKGITHG 120
121 ACDYLLKPVR IEELRNIWQH VVRRKFGNRE RNNLDFSKEC NKPQSADTDH GPYQPTCGSS 180
181 DQNGRSSRKR KELHGEDDDE GDDNDYQEND EPSAAKKPRV VWSVELHRKF VAAVNQLGID 240
241 KAVPKRILEL MNVEKLTREN VASHLQKYRL YLKRLGAVAS QQASIVAAFG GRDPSFLHIG 300
301 AFEGLQSYQP FAPSAALPSF NPHGLLTRTS AAAAFGLQEL AAPSSTIQTS TGNVTVGHCL 360
361 EENQQANLAQ GLTAAIGQPQ LQQNWIHQEG NGLSDVFSGS SLTNTLSSTL QRVPSSSLPP 420
421 QELLECKQAK VSMPPSIRIP PSSSALLERT LGVSTNLGDS SISQQGALPI DGGFSADRLP 480
481 LHSSFDGAVA TKLDTSLAAS QREIGQQGKF SVSMLVSPSD NLALAKNAKT GASSSGSTII 540
541 LPLDTARHSD YLQFGGASNS LQKMDGQKQD HIQSSNIIWS SMPSTQLPSD TQIHNTQNQR 600
601 LDSGSFNHNI GAHLADQTNA SASILPQMKF DTRISEEKMK QKNTYDLGSS KLQGGFNSSG 660
661 CNFDGLLNSI IKVEKDDLPF MDNELGCDLF PLGACI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
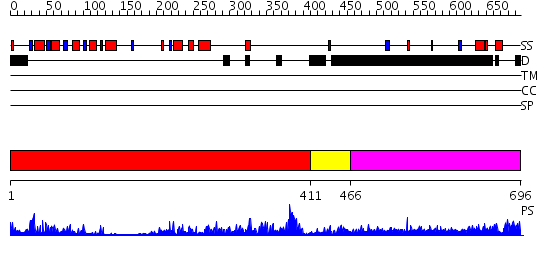
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..410] | 74.522879 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [411..465] | 77.39794 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding | |
| 3 | View Details | [466..696] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)