













| General Information: |
|
| Name(s) found: |
SCAR1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 821 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SCAR complex
[TAS]
|
| Biological Process: |
positive regulation of actin nucleation
[IMP]
|
| Molecular Function: |
actin monomer binding
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLVRLQVRN VYGLGQKELH TKVDREDPKA ILDDVAVSGL VGILRQLGDL TEFAAEIFHG 60
61 IQEEVMITAS RSNKLKMRLK QIEAKVPTIQ KTVLAQTNHI HFAYTGGLEW HPRIPNVQNH 120
121 FMYDELPPFI MTPYEDCREP PRLHLLDKFD INGPGSCLKR YSDPTHFKRA SRASKPSEIK 180
181 KKKSIQRGRD ISRLASVANQ SDRKTCTSLS FSGRTSTSKT ASTIEIESKS DLQEHRSFSF 240
241 DSRSGGEKPK RVSSSSRFTP GSRTIASVLS ESESESDSPS QDLTARGSSS VSWHEKAEIV 300
301 ECNVLQCATD EAPEVMETNF VLDAEPVSRL KEHSAVEAVQ DIKPKELEMD NEDETESEGD 360
361 DFVDALYTID SESENDEAFQ ATKEVQKNLY NDITEQETEK ISNNFSVDET KCAATSELHL 420
421 SSSPVYKSDE LIHQDPWAAS EISSGTHSYS NGFSNPLYDI SGIQEHQESE EVESSCDTES 480
481 IKTWTNGNLL GLKPSKPKII AETIPEIVED IDSETFQEHL REDYKAPFDW FTSSPPLDHM 540
541 KISFKSSETL PSSELQLKLP DEYTFSSFQL VPETIATSLP DSDSDKDTFC RSSSYISDNS 600
601 DNDNRSVSMS EQQWEEESEG IRESKRQQEL YDSFHRVNAE ASSLPVPFPK IETTNGCLVE 660
661 NVSYLQNPAE PLPPPLPPLQ WMVSKIPSAG FEDNNKQSLK DALTQAFEKN NNSLTAVKKK 720
721 EPHIVTVSDP KLVTKVHLKN NVRDYKQSHG NTNETEAGDF LHQIRTKQFN LRRVVRTKTS 780
781 SSETTMNTNI SVILEKANSI RQAVASDDGE GESDTWSDSD T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
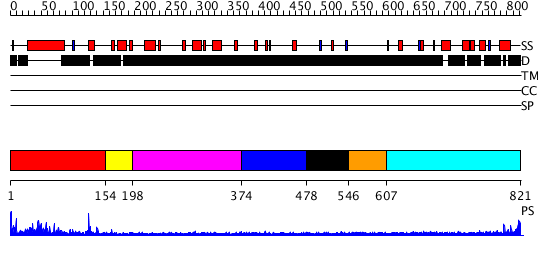
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | 2.338996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [154..197] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [198..373] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [374..477] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [478..545] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [546..606] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [607..821] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)