













| General Information: |
|
| Name(s) found: |
TELO2
[HGNC (HUGO)]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 837 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPAPSEVRL AVREAIHALS SSEDGGHIFC TLESLKRYLG EMEPPALPRE KEEFASAHFS 60
61 PVLRCLASRL SPAWLELLPH GRLEELWASF FLEGPADQAF LVLMETIEGA AGPSFRLMKM 120
121 ARLLARFLRE GRLAVLMEAQ CRQQTQPGFI LLRETLLGKV VALPDHLGNR LQQENLAEFF 180
181 PQNYFRLLGE EVVRVLQAVV DSLQGGLDSS VSFVSQVLGK ACVHGRQQEI LGVLVPRLAA 240
241 LTQGSYLHQR VCWRLVEQVP DRAMEAVLTG LVEAALGPEV LSRLLGNLVV KNKKAQFVMT 300
301 QKLLFLQSRL TTPMLQSLLG HLAMDSQRRP LLLQVLKELL ETWGSSSAIR HTPLPQQRHV 360
361 SKAVLICLAQ LGEPELRDSR DELLASMMAG VKCRLDSSLP PVRRLGMIVA EVVSARIHPE 420
421 GPPLKFQYEE DELSLELLAL ASPQPAGDGA SEAGTSLVPA TAEPPAETPA EIVDGGVPQA 480
481 QLAGSDSDLD SDDEFVPYDM SGDRELKSSK APAYVRDCVE ALTTSEDIER WEAALRALEG 540
541 LVYRSPTATR EVSVELAKVL LHLEEKTCVV GFAGLRQRAL VAVTVTDPAP VADYLTSQFY 600
601 ALNYSLRQRM DILDVLTLAA QELSRPGCLG RTPQPGSPSP NTPCLPEAAV SQPGSAVASD 660
661 WRVVVEERIR SKTQRLSKGG PRQGPAGSPS RFNSVAGHFF FPLLQRFDRP LVTFDLLGED 720
721 QLVLGRLAHT LGALMCLAVN TTVAVAMGKA LLEFVWALRF HIDAYVRQGL LSAVSSVLLS 780
781 LPAARLLEDL MDELLEARSW LADVAEKDPD EDCRTLALRA LLLLQRLKNR LLPPASP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
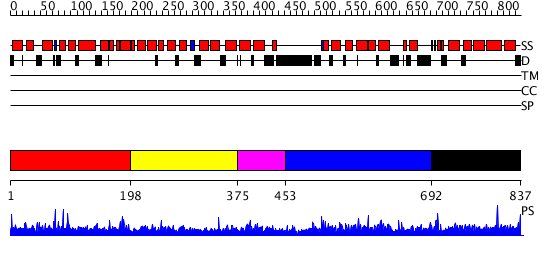
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [198..374] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [375..452] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [453..691] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [692..837] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.38 |
Source: Reynolds et al. (2008)