













| General Information: |
|
| Name(s) found: |
EF2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 843 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
membrane [IDA] cytosol [IDA] nucleolus [IDA] plasma membrane [IDA] |
| Biological Process: |
response to cold
[IMP]
|
| Molecular Function: |
copper ion binding
[IDA]
translation elongation factor activity [ISS] translation factor activity, nucleic acid binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKFTADELR RIMDYKHNIR NMSVIAHVDH GKSTLTDSLV AAAGIIAQEV AGDVRMTDTR 60
61 ADEAERGITI KSTGISLYYE MTDESLKSFT GARDGNEYLI NLIDSPGHVD FSSEVTAALR 120
121 ITDGALVVVD CIEGVCVQTE TVLRQALGER IRPVLTVNKM DRCFLELQVD GEEAYQTFSR 180
181 VIENANVIMA TYEDPLLGDV QVYPEKGTVA FSAGLHGWAF TLTNFAKMYA SKFGVVESKM 240
241 MERLWGENFF DPATRKWSGK NTGSPTCKRG FVQFCYEPIK QIIATCMNDQ KDKLWPMLAK 300
301 LGVSMKNDEK ELMGKPLMKR VMQTWLPAST ALLEMMIFHL PSPHTAQRYR VENLYEGPLD 360
361 DQYANAIRNC DPNGPLMLYV SKMIPASDKG RFFAFGRVFA GKVSTGMKVR IMGPNYIPGE 420
421 KKDLYTKSVQ RTVIWMGKRQ ETVEDVPCGN TVAMVGLDQF ITKNATLTNE KEVDAHPIRA 480
481 MKFSVSPVVR VAVQCKVASD LPKLVEGLKR LAKSDPMVVC TMEESGEHIV AGAGELHLEI 540
541 CLKDLQDDFM GGAEIIKSDP VVSFRETVCD RSTRTVMSKS PNKHNRLYME ARPMEEGLAE 600
601 AIDDGRIGPR DDPKIRSKIL AEEFGWDKDL AKKIWAFGPE TTGPNMVVDM CKGVQYLNEI 660
661 KDSVVAGFQW ASKEGPLAEE NMRGICFEVC DVVLHSDAIH RGGGQVIPTA RRVIYASQIT 720
721 AKPRLLEPVY MVEIQAPEGA LGGIYSVLNQ KRGHVFEEMQ RPGTPLYNIK AYLPVVESFG 780
781 FSSQLRAATS GQAFPQCVFD HWEMMSSDPL EPGTQASVLV ADIRKRKGLK EAMTPLSEFE 840
841 DKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
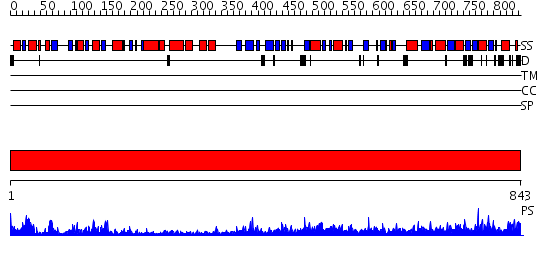
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..843] | 1000.0 | Elongation factor 2 (eEF-2), domain II; Elongation factor 2 (eEF-2), N-terminal (G) domain; Elongation factor 2 (eEF-2), domain IV; Elongation factor 2 (eEF-2) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)