













| General Information: |
|
| Name(s) found: |
UAP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 505 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA]
|
| Molecular Function: |
nucleotidyltransferase activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIEPSMEREN GALTAATTTT TAVTSPPPMA SSPRQALVER LKDYGQEDIF SLWDELSPDE 60
61 KDFLVRDIEN LDLPRIDRII RCSLHSQGLP VAAIEPVPEN WVSTVDGRTM EDREKWWKMG 120
121 LKTIYEGKLG VVLLSGGQGT RLGSSDPKGC FNIGLPSGKS LFQIQAERIL CVQRLAAQVV 180
181 SEGPIRPVTI HWYIMTSPFT DEATRKYFSS HKYFGLEPDQ ISFFQQGTLP CVTKDGKFIM 240
241 ETPFSLAKAP DGNGGVYAAL KCSRLLEDMA SRGIKYVDCY GVDNVLVRVA DPTFLGYFID 300
301 KGAASAAKVV RKAYPQEQVG VFVRRGKGGP LTVVEYSELD QSMASAINQR TGRLQYCWSN 360
361 VCLHMFTLDF LNQVATGLEK DSVYHLAEKK IPSMNGYTMG LKLEQFIFDS FPYAPSTALF 420
421 EVLREEEFAP VKNVNGSNFD TPESARLLVL RLHTRWVIAA GGFLTHSVPL YATGVEVSPL 480
481 CSYAGENLEA ICRGRTFHAP CEISL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
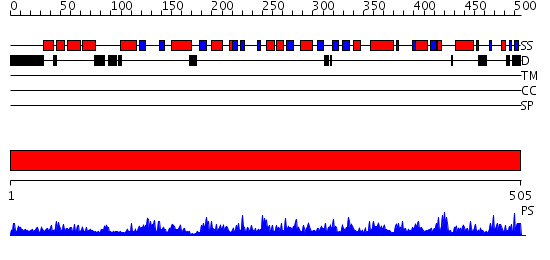
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..505] | 167.0 | UDP-N-acetylglucosamine pyrophosphorylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)