













| General Information: |
|
| Name(s) found: |
VAL1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 790 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
mitochondrion [IDA] |
| Biological Process: |
positive regulation of seed germination
[IGI]
response to abscisic acid stimulus [TAS] response to sucrose stimulus [TAS] |
| Molecular Function: |
transcription factor activity
[ISS]
transcription repressor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFEVKMGSKM CMNASCGTTS TVEWKKGWPL RSGLLADLCY RCGSAYESSL FCEQFHKDQS 60
61 GWRECYLCSK RLHCGCIASK VTIELMDYGG VGCSTCACCH QLNLNTRGEN PGVFSRLPMK 120
121 TLADRQHVNG ESGGRNEGDL FSQPLVMGGD KREEFMPHRG FGKLMSPEST TTGHRLDAAG 180
181 EMHESSPLQP SLNMGLAVNP FSPSFATEAV EGMKHISPSQ SNMVHCSASN ILQKPSRPAI 240
241 STPPVASKSA QARIGRPPVE GRGRGHLLPR YWPKYTDKEV QQISGNLNLN IVPLFEKTLS 300
301 ASDAGRIGRL VLPKACAEAY FPPISQSEGI PLKIQDVRGR EWTFQFRYWP NNNSRMYVLE 360
361 GVTPCIQSMM LQAGDTVTFS RVDPGGKLIM GSRKAANAGD MQGCGLTNGT STEDTSSSGV 420
421 TENPPSINGS SCISLIPKEL NGMPENLNSE TNGGRIGDDP TRVKEKKRTR TIGAKNKRLL 480
481 LHSEESMELR LTWEEAQDLL RPSPSVKPTI VVIEEQEIEE YDEPPVFGKR TIVTTKPSGE 540
541 QERWATCDDC SKWRRLPVDA LLSFKWTCID NVWDVSRCSC SAPEESLKEL ENVLKVGREH 600
601 KKRRTGESQA AKSQQEPCGL DALASAAVLG DTIGEPEVAT TTRHPRHRAG CSCIVCIQPP 660
661 SGKGRHKPTC GCTVCSTVKR RFKTLMMRRK KKQLERDVTA AEDKKKKDME LAESDKSKEE 720
721 KEVNTARIDL NSDPYNKEDV EAVAVEKEES RKRAIGQCSG VVAQDASDVL GVTELEGEGK 780
781 NVREEPRVSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
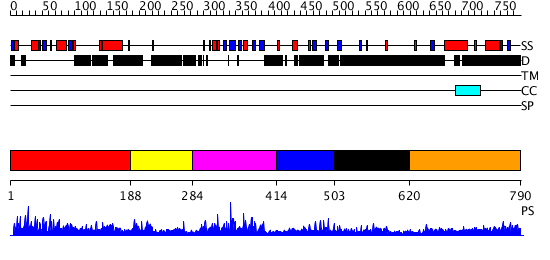
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | 1.26 | No description for 2dfyX was found. | |
| 2 | View Details | [188..283] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [284..413] | 33.30103 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 4 | View Details | [414..502] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [503..619] | 2.83 | No description for 2e61A was found. | |
| 6 | View Details | [620..790] | 3.104993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)