













| General Information: |
|
| Name(s) found: |
PGAM1_RAT
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Rattus norvegicus |
| Length: | 254 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [UNKNOWN] |
| Biological Process: |
glycolysis
[IEA]
metabolic process [IEA] regulation of glycolysis [UNKNOWN] respiratory burst [UNKNOWN] regulation of pentose-phosphate shunt [UNKNOWN] |
| Molecular Function: |
protein kinase binding
[UNKNOWN]
2,3-bisphospho-D-glycerate 2-phosphohydrolase activity [IEA] bisphosphoglycerate mutase activity [IEA] phosphoglycerate mutase activity [UNKNOWN][IDA] hydrolase activity [IEA] isomerase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAYKLVLIR HGESAWNLEN RFSGWYDADL SPAGHEEAKR GGQALRDAGY EFDICFTSVQ 60
61 KRAIRTLWTV LDAIDQMWLP VVRTWRLNER HYGGLTGLNK AETAAKHGEA QVKIWRRSYD 120
121 VPPPPMEPDH PFYSNISKDR RYADLTEDQL PSCESLKDTI ARALPFWNEE IVPQIKEGKR 180
181 VLIAAHGNSL RGIVKHLEGL SEEAIMELNL PTGIPIVYEL DKNLKPIKPM QFLGDEETVR 240
241 KAMEAVAAQG KVKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
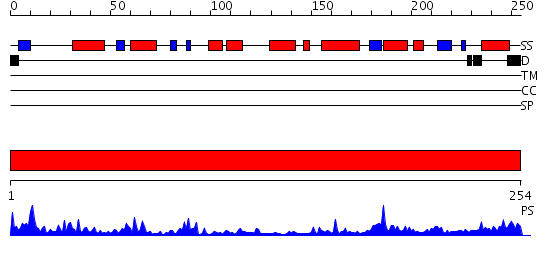
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..254] | 75.522879 | Crystal structure of human B type phosphoglycerate mutase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)