













| General Information: |
|
| Name(s) found: |
gi|29144731
[NCBI NR]
gi|29140370 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Salmonella enterica subsp. enterica serovar Typhi str. Ty2 |
| Length: | 712 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPHVMKRDG CKVPFKSERI KEAILRAAKA AGVDDADYCA TVAEVVSSQM NARSQVDINE 60
61 IQTAVENQLM SGPYKQLARA YIEYRHDRDI QREKRGRLNQ EIRGLVEQTN SALLNENANK 120
121 DSKVIPTQRD LLAGIVAKHY ARQHLLPRDV VQAHERGDIH YHDLDYSPFF PMFNCMLIDL 180
181 KGMLTQGFKM GNAEIEPPKS ISTATAVTAQ IIAQVASHIY GGTTINRIDE VLAPFVTESY 240
241 NKHRKTADEW QIPDAEGYAR SRTEKECYDA FQSLEYEVNT LHTANGQTPF VTFGFGLGTS 300
301 WESRLIQASI LRNRIAGLGK NRKTAVFPKL VFAIRDGLNH KFGDPNYDIK QLALECASKR 360
361 MYPDILNYDQ VVKVTGSFKT PMGCRSFLGV WENENGEQIH DGRNNLGVIS LNLPRIALEA 420
421 KGDETAFWKL LDERLALARK ALMTRIARLE GVKARVAPIL YMEGACGVRL KADDDVSEIF 480
481 KNGRASISLG YIGIHETINA LFGGEHLYDS EQLRAKGIAI VERLRQAVDQ WKDETGYGFS 540
541 LYSTPSENLC DRFCRLDTAE FGVVPGVTDK GYYTNSFHLD VEKKVNPYDK ISFEAPYPPL 600
601 ANGGFICYGE YPNIQHNLKA LEDVWDYSYQ HVPYYGTNTP IDECYECGFT GEFECTSKGF 660
661 TCPKCGNHDA ARVSVTRRVC GYLGSPDARP FNAGKQEEVK RRVKHLGNGQ IG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
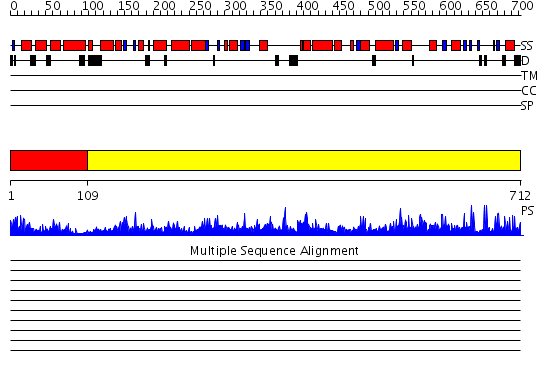
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 25.39794 | R1 subunit of ribonucleotide reductase, N-terminal domain; R1 subunit of ribonucleotide reductase, C-terminal domain | |
| 2 | View Details | [109..712] | 1000.0 | Class III anaerobic ribonucleotide reductase NRDD subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)