













| General Information: |
|
| Name(s) found: |
gi|29143103
[NCBI NR]
gi|29138736 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Salmonella enterica subsp. enterica serovar Typhi str. Ty2 |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIDTPSGVQ LRIRGKVQGV GFRPFVWQLA QQLQLHGDVC NDGDGVVVRL LEDPSQFIAA 60
61 LYQDCPPLAR IDSVEHASLV WERAPTDFTI RQSAGGSMNT QIVPDAATCP ACLAEMNTPG 120
121 ERRYRYPFIN CTHCGPRFTI IRAMPYDRPF TVMAAFPLCP ECDSEYRDPY DRRFHAQPVA 180
181 CPSCGPHLEW RSQHERAEKE AALQAAVAQL NAGGIIAVKG LGGFHLACDA RNDNAVAMLR 240
241 ARKHRPAKPL AVMLPTAQTL PTAARSLLTT PAAPIVLVDK QYVPSLSEGI APGLTEVGVM 300
301 LPANPLQHLL LQALNYPLVM TSGNLSGKPP AITNEQALDD LHDIADGFLL HNRDIVQRMD 360
361 DSVVRDSGEM LRRSRGYVPD AIALPPGFRD VPPILCLGAD LKNTFCLVRG EQAVVSQHLG 420
421 DLSDDGIQAQ WREALRLIQS IYDFTPERIV CDAHPGYVSS QWASEMRLPT ETVLHHHAHA 480
481 AACLAEHGWP LDGGEVIALT VDGIGMGENG ALWGGECLRV NYRECEHLGG LPAVALPGGD 540
541 LAAKQPWRNL LAQCLRFVPD WQNYPETAGL QQQNWSVLAR AIERGVNAPL ASSCGRLFDA 600
601 VAAALRCAPA SLSYEGEAAC ALEALASQCA NVEYPVTMPL NGAQLDVAVF WRQWLNWQAT 660
661 PAQRAWAFHD ALACGFATLM RQQATARGIT TLVFSGGVIH NRLLRARLAF YLSDFKLLFP 720
721 QRLPAGDGGL SFGQGVIAAA RALREV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
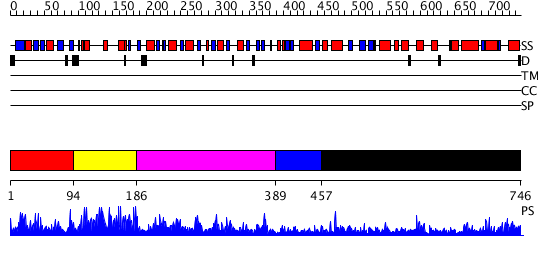
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 27.39794 | Hydrogenase maturation protein HypF N-terminal domain (HypF-ACP) | |
| 2 | View Details | [94..185] | 4.09 | No description for 2i13A was found. | |
| 3 | View Details | [186..388] | 54.69897 | Hypothetical protein YciO | |
| 4 | View Details | [389..456] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [457..746] | 50.522879 | No description for 2ivnA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)