













| General Information: |
|
| Name(s) found: |
ada-2 /
CE37767
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 583 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA intracellular [IEA |
| Biological Process: |
locomotory behavior
[IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP post-embryonic body morphogenesis [IMP gamete generation [IMP |
| Molecular Function: |
nucleic acid binding
[IEA DNA binding [IEA zinc ion binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADDTGEHPV CFNCTLRVDQ TIHVKCFECP LRICILCFQC GAESPPHRRG HNYELVKPTS 60
61 NPEAMSWTHE DEFELLKAAH KFKMGNWGEI AESIGRGRKD GHNCKDYFEK HFVRGWIGQF 120
121 SIKSSSWDRI KYGMYINQTL DSVLQKNCLE STERMLLIRD AVRASGESWD ENDPKLAIKV 180
181 QQVLEQYVQK VIDGDIETKY ERPKFLTNQY ETELSPDDCD PDNERDMKSK RSVKQEINTD 240
241 DSDNDNSPGP SSSCRKKGPP SPKRRKTTRV MRESSSDSDD DKSQASDKDT YHGMERETEE 300
301 EDPPSAFETA QEFDTEDEEE EKKPKRGTAS SSKRRKRRRM WVSKKDRRLH EFRKKMNQEV 360
361 KELRSAYPRL LHESGVSKSE TRPKMRRSDE LKILGYNNER EELECEWFNE AEQLISRLTI 420
421 SATEPQKDQR LDMENDIKFA RLRHYVRLLG IRKAKRNTVL EHDKINEFMK FYQEACYAAA 480
481 KRPVSQQEIM DSRTEKEKLL AMTQQFLTRD EYRSLRASIE RIDNVVERIE VLQDLQKNGE 540
541 TTLKNAPVER KSKKKMRKID CEQKKKIATE WSKFKKWHTE GWE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
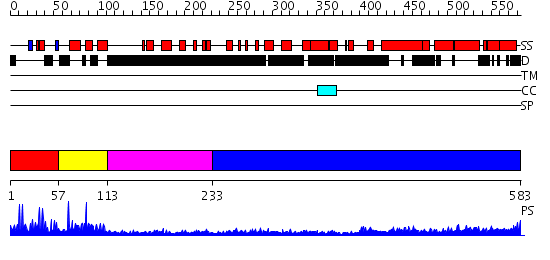
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..56] | 2.65 | No description for 2e5rA was found. | |
| 2 | View Details | [57..112] | 14.09691 | No description for 2elkA was found. | |
| 3 | View Details | [113..232] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [233..583] | 9.39794 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)