













| General Information: |
|
| Name(s) found: |
PPS_BACSU
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Bacillus subtilis subsp. subtilis str. 168 |
| Length: | 866 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLVLGLHE IEKTQLSLVG GKGLHLGELS KIQGIQVPEG FCVTTVGYQK AIEQNETLQV 60
61 LLDQLTMLKV EDRDQIGNIS RKIRQIIMEV DIPSDVVKAV AQYLSQFGEE HAYAVRSSAT 120
121 AEDLPHASFA GQQDTYLNIT GVDAILQHIS KCWASLFTDR AVIYRMQNGF DHSQVYLSVI 180
181 VQRMVFPQAS GILFTADPIT SNRKVLSIDA GFGLGEALVS GLVSADCFKV QDGQIIDKRI 240
241 ATKKMAIYGR KEGGTETQQI DSDQQKAQTL TDEQILQLAR IGRQIEAHFG QPQDIEWCLA 300
301 RDTFYIVQSR PITTLFPIPE ASDQENHVYI SVGHQQMMTD PIKPLGLSFF LLTTVAPMRK 360
361 AGGRLFVDVT HHLASPDSRE VFLKGMGQHD QLLKDALMTI IKRRDFIKSI PNDKTAPNPS 420
421 RGNADMPAQV ENDPTIVSDL IESSQTSIEE LKQNIQTKSG SDLFRFILED IQELKKILFN 480
481 PKSSVLIRTA MNASLWINEK MNEWLGEKNA ADTLSQSVPH NITSEMGLAL LDVADVIRPY 540
541 PEVIDYLQHV KDDNFLDELA KFDGGSKTRD AIYDYLSKYG MRCTGEIDIT RTRWSEKPTT 600
601 LVPMILNNIK NFEPNVGHRK FEQGRQEALK KEQELLDRLK QLPDGEQKAK ETKRAIDIIR 660
661 NFSGFREYPK YGMVNRYFVY KQALLKEAEQ LIEAGVIHEK EDIYYLTFEE LHEVVRTHKL 720
721 DYQIISTRKD EYTLYEKLSP PRVITSDGEI VTGEYKRENL PAGAIVGLPV SSGVIEGRAR 780
781 VILNMEDADL EDGDILVTSF TDPSWTPLFV SIKGLVTEVG GLMTHGAVIA REYGLPAVVG 840
841 VENAAKLIKD GQRIRVHGTE GYIEIF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
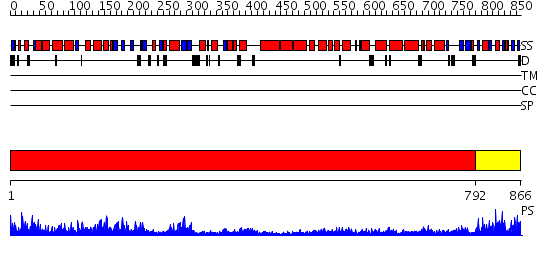
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..791] | 169.0 | No description for 2olsA was found. | |
| 2 | View Details | [792..866] | 32.69897 | Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain; N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)