













| General Information: |
|
| Name(s) found: |
gi|34897354
[NCBI NR]
gi|15042816 [NCBI NR] gi|115456321 [NCBI NR] gi|113550232 [NCBI NR] gi|108711847 [NCBI NR] gi|108711846 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQISLEHRY KECRPRRNNS CPTKTLVGKD QLKELEHRRP SPSVIAKLMG LDVLPPAYVA 60
61 HNQHQEFKDV FEVSEEPQEA VTKERSHNFP KGLPSLKRSA LKLRKLMPSK SPYGDETFDN 120
121 NVVNQDGFDR LNSLEINNPL FEKHPYDVNC SPNYRYEKDS TSSTFRKYPV GLGNSSLKEI 180
181 VVLELGLGEV QHSGNAFSTP EPSDVNKNFR RKMKQAEFST TNRGSQNLLG TKDINVPRIK 240
241 GERHLTSNAV DSLLKRQDSS LDQYNTVDTD NTGSSQKCVS SEVNSRKSNR SSSNSSPWKI 300
301 RRKYEEGAIG SKTLAEMFAL SDSERLKRDS DSHVQIQDNK LNRGNNNDKE GCFIVLPKHA 360
361 PRLPPHSLLD KNSSCERSPH DIFFSNTSIS HNSGQFHFDS FWDKPTRQQI SSPTQDDLRN 420
421 ASCARYHTLE QHRSSFPSYD NTRNNSWHLT DDFSTFACIN EKVLFTTDED LLRKPTETVH 480
481 SSFGSRLSGE QKVSASPFHC GVYEAITISD HTCAAKSRRS LKEVDRPSPV SILEPPTDED 540
541 SCCSGYLKND SQVMPSIDKQ IYGCELRYEQ EVSLSSDNDN DSSDQSLEAF EVEEEKEFSY 600
601 LLDILISSGV IVADSQLLFK SWQSSGYLVG PHVFDKLEGK YSKVATWPRP QRRLLFDLAN 660
661 SVLSEILAPC IDTHPWAKLS RNCCPVWGPE GPVEVVWQTM VRQQEELAVA HPDDKILDPE 720
721 WLEFGEGINM VGWHIARMLH GDLLDDVILE FLSGFVAS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
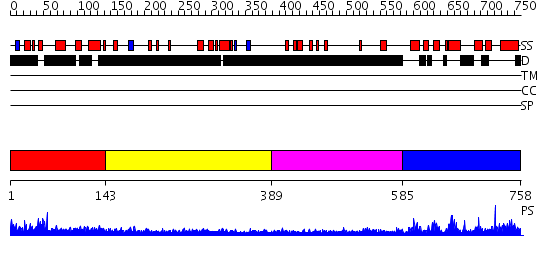
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [143..388] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [389..584] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [585..758] | 3.059984 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)