













| General Information: |
|
| Name(s) found: |
gi|51964060
[NCBI NR]
gi|50908867 [NCBI NR] gi|48716973 [NCBI NR] gi|125582253 [NCBI NR] gi|115446299 [NCBI NR] gi|113536460 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 734 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVELNSRAL MDEALKARDA AERKFHARDV KGARRSAIKA QNLCPSLDGI SQMVSTLEVL 60
61 LASESKVDGE NDWYRILSLS ASADEEEVKK QYRKLALQLH PDKNKSVGAE GAFKLISEAW 120
121 AVLSDKSRKM QYDQKRKDHP VTNGANGLYT YDKKAHKRAR KNAAASAAAA AAAAAAAAEA 180
181 TTRPVGLDTF WTSCNRCRMQ YEYLRIYLNH NLLCPNCHHA FMAVETGYPC NGTSSSFSWS 240
241 TKQQQQNHKH SYSSASRTSG VPGTGHGVYQ QENTYETYNN QSFQWNQYSK TNNSAGTNAY 300
301 SSTASEKPKR KHEESYIYNY SSSGNEFGQE RPTSGRGRFS KRRQNINNGY ASVDCNGDNK 360
361 ETVAATAGTT VLADVGRVNG TSVEKFRSAV SGRRANVMRE IFQLDTRGLL IEKAKAAIRE 420
421 KLQDLNISAT RHIAAKGKAE RKNHVDHDVK GNGILPHNPS HKFKICNSKG ADVENPATDE 480
481 NNLEQKRVPV SIDVPDPDFY DFDKDRTERT FDNDQVWATY DSEDGMPRLY AMVQKVISRK 540
541 PFRIRMSFLN SKSNIELSPI NWVASGFSKT CGDFRVGRYQ IFETVNIFSH RVSWSKGPRG 600
601 IIKIVPKKGD TWALYRNWSS DWNELTPDDV IYKYEIVEVI DDFTDEQGVT VIPLLKVAGF 660
661 KAVFHRRTDS DVVRRIPKEE LFRFSHRVPS RLLTGEEGNN APKGCHELDP AATPVDLLKV 720
721 ITEVKEVATT EISE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
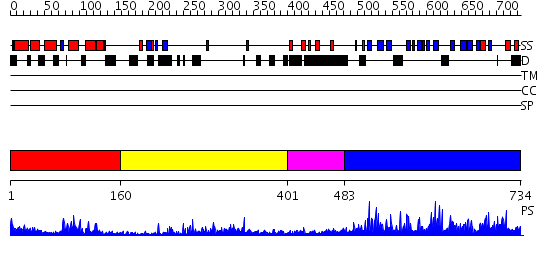
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | 17.69897 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [160..400] | 31.0 | The crystal structure of Hsp40 Ydj1 | |
| 3 | View Details | [401..482] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [483..734] | 3.03495 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)