













| General Information: |
|
| Name(s) found: |
gi|37533920
[NCBI NR]
gi|31431830 [NCBI NR] gi|215695184 [NCBI NR] gi|14018055 [NCBI NR] gi|115481820 [NCBI NR] gi|113639112 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 839 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVCFGSKALR RKGDKQRCLP EGDIDDSLPM RRGRKDKSEK PRKGGSSSSN RPSAEKAKHG 60
61 RKKSGDGKKS KGRGKSCHSD SSIEMNPGHM KNDNTLLPSK ASKPVTNFLR KRVDPETAKY 120
121 FLEISYLFDN KEIDLEERAT ICANALEETK GKELELATDG PISHTLQVLV EGCELEQLCV 180
181 FLHNSIESFH IIAVDKFGSH VAEAALKSLA THLEDEASRG IIEDILIRIC KVIAGDAANV 240
241 MSSCYGSHVL RTLLCLCKGV PLQSLQDFHT TKRSAVLAER LSCGSTRSGG SDPKNQGCGF 300
301 SDIFKSFVRE MLQNAKDDIA SLETDKNSSL VLQTALKLSA GDDHELNYII SILLGFDEDD 360
361 TAQKKDRSEQ KNEIIALLED TAYSHLLEVI VEVAPEELRN NMLTGTLKGA LFAISSHHCG 420
421 NYVVQALVAS AKTSDQMEQI WDELGSRINE LLELGKAGVV ASILAACQRL ETKRLESSQV 480
481 LSAALSSNSE SSDSIVAHML FLENYLHQKS SWEWPLGAKM SVLGCLMLQS ILQYPHQYIR 540
541 PYVASLLAMD DNKILQISKD SGGSRVLEAF LCSSATAKRK FKVFAKLQGH YGEIAMNPSG 600
601 SFLVEKCFTA SNFSHKEAIV VELLAVQTEL SRTRHGFHLL KKLDVDRYSR RPDQWRASQT 660
661 SKETTQREFQ VEFGLSSKGA GQNIEELLTS RSPSKKRKQK DKTDVTTEDA STNKQDLSHV 720
721 GKTKRIKSEK TTCEKESSNK KPTNEDSGTS MAFLKNSAKR KSPGFLSDKP SFKRQKHHKP 780
781 NAGNSSGKMF VRDSAGTPFV RNSGKQKRSI AELADLAGKE KLSASEVRKL LKTEMPGKS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
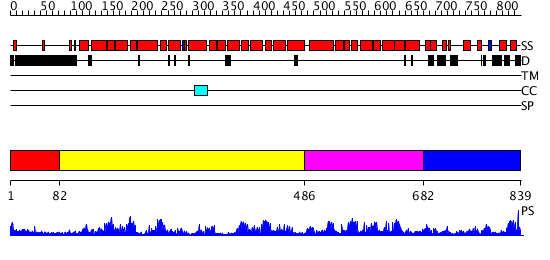
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [82..485] | 82.39794 | Pumilio 1 | |
| 3 | View Details | [486..681] | 2.108969 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [682..839] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)