













| General Information: |
|
| Name(s) found: |
gi|37532658
[NCBI NR]
gi|31431087 [NCBI NR] gi|22725930 [NCBI NR] gi|19881543 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 823 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSASPSVA PSAEYAEHLS NLVATPSLSH EHHYFLGSIG NLDPTEFIIA ETNRIPFRLA 60
61 NPNLGHWKNT FKSWPSLDKT SPEKSWAMWY KRVSASKQVH WDELGIGQAL ALTLANSAKD 120
121 EPLMASAAYF WSNTLKAFLF NQGPMTPTLL DITMITGLDV TSSANPMSLN TKNQFDFRTK 180
181 SIGGWSGYVA AYMGKGSVTP REHTAFLLMW LEKFLFCGSS CGLTTNWQFL AEALETKRQF 240
241 PLGKILLDYL YQMLSNASAK IVTGSVLSAG CESATFDRRR IPKAGADHSF YEGFQRDARV 300
301 WFPYEDSVNF DLPADFKFED INSEKFQKSR EVFSAAISPC ILPVGIHQGR NIQVSYEFYH 360
361 PMSSARKSGM GQLPIGLFFT DKIQCSGEIS STLMMDLLLN IPGPPLASIE NIELARFRSK 420
421 NFDRWWGEWK QHIFHQSASI YMTDLFPDVV PQTTKSSPPR TSNRGKDIEY APGLLPNGGR 480
481 KRKTKASAID PSALAPKKKT KKNKPKPADD LPTLDPSIEQ ALDEEEIGED FDQAAAELSD 540
541 AGEKTPSASP KQTPPNPAAP AHFSRKKKTA VKKKSATTTL KPAPPQMSSD RTPSATASHN 600
601 IEEEEQPTAP AIPVLADLFS FDIKDYFDEE AEEETTSKAL APLDETFKKT LEDISLRLES 660
661 SLDNLVADYG SIRARFAKIQ ALIPDDLANT ITPAVYLEQH QFKLEKAKQR IADRRERKDI 720
721 KATIQANRQL SNIDWLEARK IDLIAQLEEC NAELDLEKQK LANLPNAVEE QKSRLKSAIK 780
781 NVADMTKSFK AIFATDAQDI QAIEEVDQIR QRAVSAIQRY LSA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
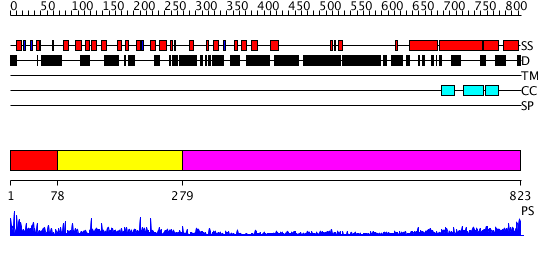
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 1.103989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [78..278] | 10.259637 | No description for PF10536.1 was found. No confident structure predictions are available. | |
| 3 | View Details | [279..823] | 6.69897 | No description for 2dfsA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)